
Selaginella moellendorffii (Spikemoss)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Lycopodiopsida; Selaginellales; Selaginellaceae; Selaginella
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
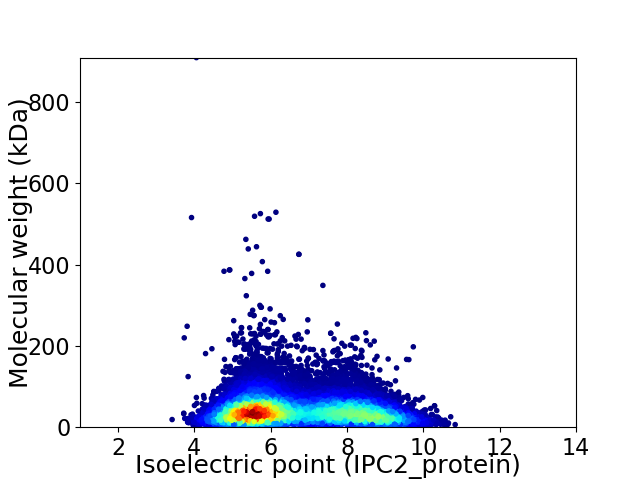
Virtual 2D-PAGE plot for 33150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8SB95|D8SB95_SELML TPR_REGION domain-containing protein OS=Selaginella moellendorffii OX=88036 GN=SELMODRAFT_420200 PE=4 SV=1
MM1 pKa = 7.18LVSVEE6 pKa = 4.29SQSQSEE12 pKa = 4.4SATCKK17 pKa = 8.28EE18 pKa = 3.99QEE20 pKa = 4.09MLLLVNFKK28 pKa = 10.99AGFTDD33 pKa = 3.59SQNMLVHH40 pKa = 6.85WNQNNTNCCKK50 pKa = 10.33WNGITCDD57 pKa = 3.87SLQEE61 pKa = 4.17MIITTAPYY69 pKa = 10.27INGPLPSEE77 pKa = 4.04LAGLTTLQTLIITGTTVWGSIPSEE101 pKa = 4.16LGNLPQLRR109 pKa = 11.84VLDD112 pKa = 4.61LSSNMLSGSIPRR124 pKa = 11.84NLGRR128 pKa = 11.84LQTLRR133 pKa = 11.84EE134 pKa = 4.04LQLASNNLSGSIPWEE149 pKa = 3.98LGSIRR154 pKa = 11.84RR155 pKa = 11.84AYY157 pKa = 10.27LVNLSNNSLSGQIPDD172 pKa = 3.71SLANIAPSGSIDD184 pKa = 3.63LSNNLFTGRR193 pKa = 11.84FPTALCRR200 pKa = 11.84LEE202 pKa = 4.09NRR204 pKa = 11.84SFLFYY209 pKa = 11.1LDD211 pKa = 4.32LSEE214 pKa = 4.57NQLSGALPASLPTTTGSHH232 pKa = 5.08EE233 pKa = 4.37VYY235 pKa = 10.37SYY237 pKa = 11.89LSFLSLASNSLTGTIPSALWSNLSSLTAVDD267 pKa = 4.64FSNNHH272 pKa = 5.72FSGEE276 pKa = 4.15IPTEE280 pKa = 3.76LAGLVGLTSLNLSRR294 pKa = 11.84NDD296 pKa = 3.49LSGEE300 pKa = 3.86IPTSISNGNALQLIDD315 pKa = 5.08LSRR318 pKa = 11.84NTLNGTIPPEE328 pKa = 5.47IGDD331 pKa = 3.98LYY333 pKa = 9.68MLEE336 pKa = 4.29MLDD339 pKa = 4.26LSYY342 pKa = 11.37NQLSGSIPTALDD354 pKa = 3.73DD355 pKa = 4.87LLSLAAFNEE364 pKa = 4.16IYY366 pKa = 10.48LYY368 pKa = 11.25SNNLNGSIPDD378 pKa = 4.7AIANLTRR385 pKa = 11.84LATLDD390 pKa = 3.83LSSNHH395 pKa = 6.99LDD397 pKa = 3.7GQIPGPAIAQLTGLQVMDD415 pKa = 5.2LSANDD420 pKa = 3.53LTGNIPSEE428 pKa = 4.0LADD431 pKa = 4.22LGQLATLDD439 pKa = 4.21LSWNQLSGVIPPEE452 pKa = 3.65IHH454 pKa = 7.13DD455 pKa = 4.61LSSLEE460 pKa = 4.07YY461 pKa = 10.58FSVANNNLSGPIPAEE476 pKa = 4.04LGSFDD481 pKa = 4.35ASSFEE486 pKa = 5.26DD487 pKa = 3.54NAGLCGFPLDD497 pKa = 4.33PCSSS501 pKa = 3.51
MM1 pKa = 7.18LVSVEE6 pKa = 4.29SQSQSEE12 pKa = 4.4SATCKK17 pKa = 8.28EE18 pKa = 3.99QEE20 pKa = 4.09MLLLVNFKK28 pKa = 10.99AGFTDD33 pKa = 3.59SQNMLVHH40 pKa = 6.85WNQNNTNCCKK50 pKa = 10.33WNGITCDD57 pKa = 3.87SLQEE61 pKa = 4.17MIITTAPYY69 pKa = 10.27INGPLPSEE77 pKa = 4.04LAGLTTLQTLIITGTTVWGSIPSEE101 pKa = 4.16LGNLPQLRR109 pKa = 11.84VLDD112 pKa = 4.61LSSNMLSGSIPRR124 pKa = 11.84NLGRR128 pKa = 11.84LQTLRR133 pKa = 11.84EE134 pKa = 4.04LQLASNNLSGSIPWEE149 pKa = 3.98LGSIRR154 pKa = 11.84RR155 pKa = 11.84AYY157 pKa = 10.27LVNLSNNSLSGQIPDD172 pKa = 3.71SLANIAPSGSIDD184 pKa = 3.63LSNNLFTGRR193 pKa = 11.84FPTALCRR200 pKa = 11.84LEE202 pKa = 4.09NRR204 pKa = 11.84SFLFYY209 pKa = 11.1LDD211 pKa = 4.32LSEE214 pKa = 4.57NQLSGALPASLPTTTGSHH232 pKa = 5.08EE233 pKa = 4.37VYY235 pKa = 10.37SYY237 pKa = 11.89LSFLSLASNSLTGTIPSALWSNLSSLTAVDD267 pKa = 4.64FSNNHH272 pKa = 5.72FSGEE276 pKa = 4.15IPTEE280 pKa = 3.76LAGLVGLTSLNLSRR294 pKa = 11.84NDD296 pKa = 3.49LSGEE300 pKa = 3.86IPTSISNGNALQLIDD315 pKa = 5.08LSRR318 pKa = 11.84NTLNGTIPPEE328 pKa = 5.47IGDD331 pKa = 3.98LYY333 pKa = 9.68MLEE336 pKa = 4.29MLDD339 pKa = 4.26LSYY342 pKa = 11.37NQLSGSIPTALDD354 pKa = 3.73DD355 pKa = 4.87LLSLAAFNEE364 pKa = 4.16IYY366 pKa = 10.48LYY368 pKa = 11.25SNNLNGSIPDD378 pKa = 4.7AIANLTRR385 pKa = 11.84LATLDD390 pKa = 3.83LSSNHH395 pKa = 6.99LDD397 pKa = 3.7GQIPGPAIAQLTGLQVMDD415 pKa = 5.2LSANDD420 pKa = 3.53LTGNIPSEE428 pKa = 4.0LADD431 pKa = 4.22LGQLATLDD439 pKa = 4.21LSWNQLSGVIPPEE452 pKa = 3.65IHH454 pKa = 7.13DD455 pKa = 4.61LSSLEE460 pKa = 4.07YY461 pKa = 10.58FSVANNNLSGPIPAEE476 pKa = 4.04LGSFDD481 pKa = 4.35ASSFEE486 pKa = 5.26DD487 pKa = 3.54NAGLCGFPLDD497 pKa = 4.33PCSSS501 pKa = 3.51
Molecular weight: 53.51 kDa
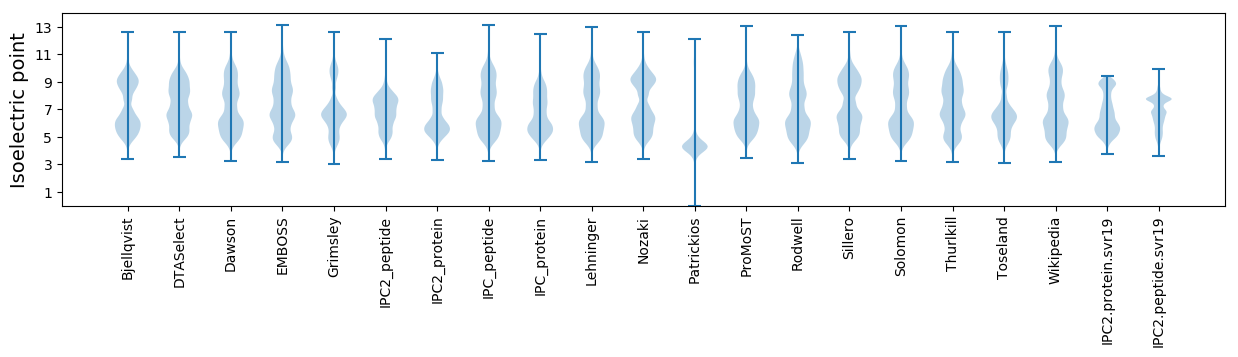
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8S3Z4|D8S3Z4_SELML Uncharacterized protein OS=Selaginella moellendorffii OX=88036 GN=SELMODRAFT_108395 PE=3 SV=1
MM1 pKa = 7.87PSHH4 pKa = 5.64KK5 pKa = 9.39TFRR8 pKa = 11.84IKK10 pKa = 10.58QKK12 pKa = 10.3LGKK15 pKa = 9.75KK16 pKa = 9.39LRR18 pKa = 11.84QNRR21 pKa = 11.84PIPHH25 pKa = 7.75WIRR28 pKa = 11.84MRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
MM1 pKa = 7.87PSHH4 pKa = 5.64KK5 pKa = 9.39TFRR8 pKa = 11.84IKK10 pKa = 10.58QKK12 pKa = 10.3LGKK15 pKa = 9.75KK16 pKa = 9.39LRR18 pKa = 11.84QNRR21 pKa = 11.84PIPHH25 pKa = 7.75WIRR28 pKa = 11.84MRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
Molecular weight: 6.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13328660 |
30 |
8462 |
402.1 |
44.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.417 ± 0.013 | 1.905 ± 0.007 |
5.273 ± 0.008 | 6.429 ± 0.018 |
4.096 ± 0.009 | 6.847 ± 0.014 |
2.402 ± 0.006 | 4.705 ± 0.01 |
5.351 ± 0.013 | 10.132 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.006 | 3.437 ± 0.006 |
4.826 ± 0.013 | 3.817 ± 0.009 |
6.002 ± 0.011 | 8.13 ± 0.014 |
4.775 ± 0.008 | 6.977 ± 0.012 |
1.454 ± 0.005 | 2.669 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
