
Wenzhou tombus-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.54
Get precalculated fractions of proteins
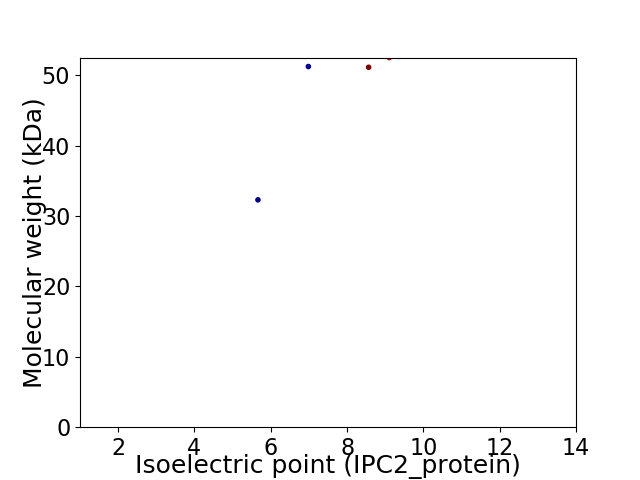
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH00|A0A1L3KH00_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 12 OX=1923665 PE=4 SV=1
MM1 pKa = 7.98EE2 pKa = 5.65DD3 pKa = 3.18QSSDD7 pKa = 3.51DD8 pKa = 3.67FEE10 pKa = 6.03SIVVSNSEE18 pKa = 3.74SSVRR22 pKa = 11.84IGSTEE27 pKa = 3.62PRR29 pKa = 11.84RR30 pKa = 11.84CGIDD34 pKa = 3.14PGDD37 pKa = 4.87SISQRR42 pKa = 11.84DD43 pKa = 3.67PGEE46 pKa = 4.18SSSEE50 pKa = 4.06EE51 pKa = 3.91EE52 pKa = 4.28EE53 pKa = 4.27EE54 pKa = 4.14EE55 pKa = 5.17QFQEE59 pKa = 4.42DD60 pKa = 4.63CGSSTSSCGSSSTSHH75 pKa = 6.57SSSQRR80 pKa = 11.84GSNSGGTVGGRR91 pKa = 11.84RR92 pKa = 11.84EE93 pKa = 4.41SGLLMAPGDD102 pKa = 3.56AGGEE106 pKa = 4.05VRR108 pKa = 11.84EE109 pKa = 4.46SVNQLEE115 pKa = 4.37EE116 pKa = 3.99HH117 pKa = 6.89DD118 pKa = 4.82CPFKK122 pKa = 11.02HH123 pKa = 6.55EE124 pKa = 4.25RR125 pKa = 11.84TTLFNPNYY133 pKa = 9.55NCCDD137 pKa = 3.16LGAWVRR143 pKa = 11.84PSVVEE148 pKa = 3.8YY149 pKa = 10.57DD150 pKa = 3.85KK151 pKa = 11.27PWFLFWSKK159 pKa = 10.65RR160 pKa = 11.84RR161 pKa = 11.84DD162 pKa = 3.58LVFSPVLVDD171 pKa = 3.33YY172 pKa = 8.86LKK174 pKa = 10.94PMAMLRR180 pKa = 11.84EE181 pKa = 4.04RR182 pKa = 11.84DD183 pKa = 3.62NALGLSLLTAARR195 pKa = 11.84NKK197 pKa = 9.47TISALGIHH205 pKa = 6.35PTWITPIFPGSISMAMQVGPMEE227 pKa = 4.34KK228 pKa = 9.42RR229 pKa = 11.84AKK231 pKa = 9.81EE232 pKa = 3.87LQKK235 pKa = 10.79PLIKK239 pKa = 10.59GKK241 pKa = 10.74LPGNWTLRR249 pKa = 11.84STHH252 pKa = 6.42KK253 pKa = 10.29PLEE256 pKa = 4.09RR257 pKa = 11.84LVEE260 pKa = 4.18NNSRR264 pKa = 11.84LGMSRR269 pKa = 11.84VRR271 pKa = 11.84SNQFVRR277 pKa = 11.84KK278 pKa = 8.77WIFGQSPEE286 pKa = 4.33LPRR289 pKa = 11.84IGG291 pKa = 3.91
MM1 pKa = 7.98EE2 pKa = 5.65DD3 pKa = 3.18QSSDD7 pKa = 3.51DD8 pKa = 3.67FEE10 pKa = 6.03SIVVSNSEE18 pKa = 3.74SSVRR22 pKa = 11.84IGSTEE27 pKa = 3.62PRR29 pKa = 11.84RR30 pKa = 11.84CGIDD34 pKa = 3.14PGDD37 pKa = 4.87SISQRR42 pKa = 11.84DD43 pKa = 3.67PGEE46 pKa = 4.18SSSEE50 pKa = 4.06EE51 pKa = 3.91EE52 pKa = 4.28EE53 pKa = 4.27EE54 pKa = 4.14EE55 pKa = 5.17QFQEE59 pKa = 4.42DD60 pKa = 4.63CGSSTSSCGSSSTSHH75 pKa = 6.57SSSQRR80 pKa = 11.84GSNSGGTVGGRR91 pKa = 11.84RR92 pKa = 11.84EE93 pKa = 4.41SGLLMAPGDD102 pKa = 3.56AGGEE106 pKa = 4.05VRR108 pKa = 11.84EE109 pKa = 4.46SVNQLEE115 pKa = 4.37EE116 pKa = 3.99HH117 pKa = 6.89DD118 pKa = 4.82CPFKK122 pKa = 11.02HH123 pKa = 6.55EE124 pKa = 4.25RR125 pKa = 11.84TTLFNPNYY133 pKa = 9.55NCCDD137 pKa = 3.16LGAWVRR143 pKa = 11.84PSVVEE148 pKa = 3.8YY149 pKa = 10.57DD150 pKa = 3.85KK151 pKa = 11.27PWFLFWSKK159 pKa = 10.65RR160 pKa = 11.84RR161 pKa = 11.84DD162 pKa = 3.58LVFSPVLVDD171 pKa = 3.33YY172 pKa = 8.86LKK174 pKa = 10.94PMAMLRR180 pKa = 11.84EE181 pKa = 4.04RR182 pKa = 11.84DD183 pKa = 3.62NALGLSLLTAARR195 pKa = 11.84NKK197 pKa = 9.47TISALGIHH205 pKa = 6.35PTWITPIFPGSISMAMQVGPMEE227 pKa = 4.34KK228 pKa = 9.42RR229 pKa = 11.84AKK231 pKa = 9.81EE232 pKa = 3.87LQKK235 pKa = 10.79PLIKK239 pKa = 10.59GKK241 pKa = 10.74LPGNWTLRR249 pKa = 11.84STHH252 pKa = 6.42KK253 pKa = 10.29PLEE256 pKa = 4.09RR257 pKa = 11.84LVEE260 pKa = 4.18NNSRR264 pKa = 11.84LGMSRR269 pKa = 11.84VRR271 pKa = 11.84SNQFVRR277 pKa = 11.84KK278 pKa = 8.77WIFGQSPEE286 pKa = 4.33LPRR289 pKa = 11.84IGG291 pKa = 3.91
Molecular weight: 32.3 kDa
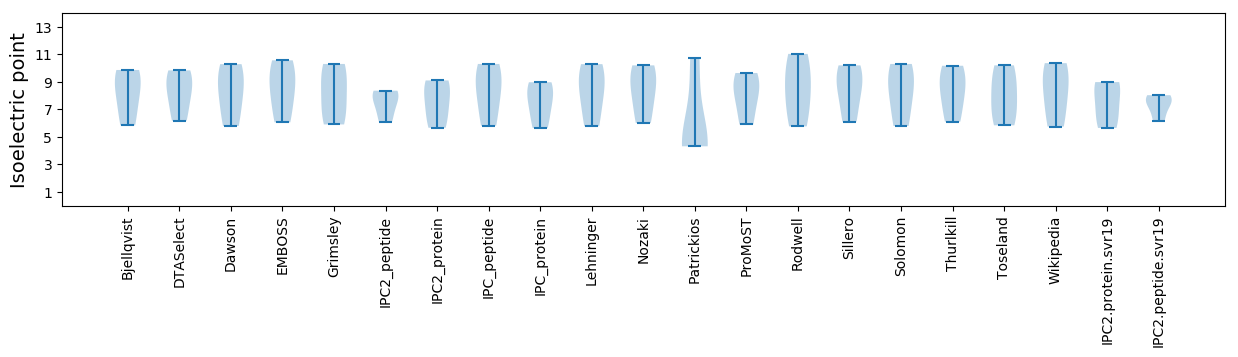
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH00|A0A1L3KH00_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 12 OX=1923665 PE=4 SV=1
MM1 pKa = 7.93RR2 pKa = 11.84AQKK5 pKa = 10.18SANAVGGAVRR15 pKa = 11.84TPIKK19 pKa = 10.21GSKK22 pKa = 9.06PRR24 pKa = 11.84TSAEE28 pKa = 3.92VQKK31 pKa = 9.59TCRR34 pKa = 11.84QCKK37 pKa = 9.88APTRR41 pKa = 11.84LVKK44 pKa = 10.8GFVVCQQDD52 pKa = 3.46PNHH55 pKa = 6.05NFKK58 pKa = 10.23PVKK61 pKa = 9.49IVCGMCGAGRR71 pKa = 11.84SDD73 pKa = 3.52LVIYY77 pKa = 9.11PGKK80 pKa = 10.05VWCTKK85 pKa = 10.22CAATTIDD92 pKa = 3.46KK93 pKa = 9.94QAKK96 pKa = 8.38VEE98 pKa = 4.18KK99 pKa = 10.34KK100 pKa = 9.19VVNNKK105 pKa = 8.27VVQPKK110 pKa = 10.06AGKK113 pKa = 9.35PKK115 pKa = 9.04VKK117 pKa = 9.98SVKK120 pKa = 9.93SVPGNSPQKK129 pKa = 10.6KK130 pKa = 9.51KK131 pKa = 10.82IPFQPIGVNSRR142 pKa = 11.84GLKK145 pKa = 10.52SGDD148 pKa = 3.45LKK150 pKa = 11.02WFGKK154 pKa = 8.52TKK156 pKa = 7.57QTKK159 pKa = 4.59TTKK162 pKa = 10.34PKK164 pKa = 10.18FAWIKK169 pKa = 9.03KK170 pKa = 7.1TVGNVKK176 pKa = 10.22GLEE179 pKa = 3.82SHH181 pKa = 7.24IFYY184 pKa = 10.29TNKK187 pKa = 9.12TMQGAQLAALRR198 pKa = 11.84EE199 pKa = 4.51SPCFSCGGKK208 pKa = 9.66FATGGGWSSIEE219 pKa = 4.17PPSSSGKK226 pKa = 9.26ISKK229 pKa = 10.61GSIITMACPRR239 pKa = 11.84NTCLSEE245 pKa = 4.16YY246 pKa = 10.04NVVVVVEE253 pKa = 4.37KK254 pKa = 10.63KK255 pKa = 10.26KK256 pKa = 11.05KK257 pKa = 8.63EE258 pKa = 4.2FKK260 pKa = 9.94TLPLTGKK267 pKa = 9.5GKK269 pKa = 10.66KK270 pKa = 9.25ILTPQPPLAPEE281 pKa = 3.96TRR283 pKa = 11.84PVDD286 pKa = 3.9LSEE289 pKa = 4.95DD290 pKa = 3.54QVKK293 pKa = 8.03TVFDD297 pKa = 4.27EE298 pKa = 4.38EE299 pKa = 4.68SSRR302 pKa = 11.84HH303 pKa = 4.64NLRR306 pKa = 11.84EE307 pKa = 4.13KK308 pKa = 10.37IVSAFVAANEE318 pKa = 4.1DD319 pKa = 3.21FRR321 pKa = 11.84LADD324 pKa = 3.27SGARR328 pKa = 11.84QTLVNNARR336 pKa = 11.84RR337 pKa = 11.84QIKK340 pKa = 10.01LLEE343 pKa = 4.09RR344 pKa = 11.84QMPQRR349 pKa = 11.84LNLRR353 pKa = 11.84QKK355 pKa = 8.12TGPVRR360 pKa = 11.84TVPLRR365 pKa = 11.84WKK367 pKa = 9.96INRR370 pKa = 11.84RR371 pKa = 11.84TILNPLWSVTLSLLSEE387 pKa = 4.31SDD389 pKa = 3.26QLNPEE394 pKa = 4.13DD395 pKa = 4.13VEE397 pKa = 5.12LIQEE401 pKa = 4.29TAYY404 pKa = 10.68HH405 pKa = 5.8SVILEE410 pKa = 3.9SLLPKK415 pKa = 10.15KK416 pKa = 10.22KK417 pKa = 10.26KK418 pKa = 10.25KK419 pKa = 10.4NNSKK423 pKa = 8.5KK424 pKa = 8.06TAEE427 pKa = 4.25AVHH430 pKa = 5.55QVAEE434 pKa = 4.31AAPPPTPHH442 pKa = 7.18PSEE445 pKa = 3.95EE446 pKa = 4.2VTAEE450 pKa = 4.11EE451 pKa = 4.64LWGDD455 pKa = 3.87DD456 pKa = 4.43ASPDD460 pKa = 3.9SSWLQEE466 pKa = 4.27TQEE469 pKa = 4.03EE470 pKa = 5.11RR471 pKa = 11.84YY472 pKa = 10.14EE473 pKa = 4.13NQQ475 pKa = 2.85
MM1 pKa = 7.93RR2 pKa = 11.84AQKK5 pKa = 10.18SANAVGGAVRR15 pKa = 11.84TPIKK19 pKa = 10.21GSKK22 pKa = 9.06PRR24 pKa = 11.84TSAEE28 pKa = 3.92VQKK31 pKa = 9.59TCRR34 pKa = 11.84QCKK37 pKa = 9.88APTRR41 pKa = 11.84LVKK44 pKa = 10.8GFVVCQQDD52 pKa = 3.46PNHH55 pKa = 6.05NFKK58 pKa = 10.23PVKK61 pKa = 9.49IVCGMCGAGRR71 pKa = 11.84SDD73 pKa = 3.52LVIYY77 pKa = 9.11PGKK80 pKa = 10.05VWCTKK85 pKa = 10.22CAATTIDD92 pKa = 3.46KK93 pKa = 9.94QAKK96 pKa = 8.38VEE98 pKa = 4.18KK99 pKa = 10.34KK100 pKa = 9.19VVNNKK105 pKa = 8.27VVQPKK110 pKa = 10.06AGKK113 pKa = 9.35PKK115 pKa = 9.04VKK117 pKa = 9.98SVKK120 pKa = 9.93SVPGNSPQKK129 pKa = 10.6KK130 pKa = 9.51KK131 pKa = 10.82IPFQPIGVNSRR142 pKa = 11.84GLKK145 pKa = 10.52SGDD148 pKa = 3.45LKK150 pKa = 11.02WFGKK154 pKa = 8.52TKK156 pKa = 7.57QTKK159 pKa = 4.59TTKK162 pKa = 10.34PKK164 pKa = 10.18FAWIKK169 pKa = 9.03KK170 pKa = 7.1TVGNVKK176 pKa = 10.22GLEE179 pKa = 3.82SHH181 pKa = 7.24IFYY184 pKa = 10.29TNKK187 pKa = 9.12TMQGAQLAALRR198 pKa = 11.84EE199 pKa = 4.51SPCFSCGGKK208 pKa = 9.66FATGGGWSSIEE219 pKa = 4.17PPSSSGKK226 pKa = 9.26ISKK229 pKa = 10.61GSIITMACPRR239 pKa = 11.84NTCLSEE245 pKa = 4.16YY246 pKa = 10.04NVVVVVEE253 pKa = 4.37KK254 pKa = 10.63KK255 pKa = 10.26KK256 pKa = 11.05KK257 pKa = 8.63EE258 pKa = 4.2FKK260 pKa = 9.94TLPLTGKK267 pKa = 9.5GKK269 pKa = 10.66KK270 pKa = 9.25ILTPQPPLAPEE281 pKa = 3.96TRR283 pKa = 11.84PVDD286 pKa = 3.9LSEE289 pKa = 4.95DD290 pKa = 3.54QVKK293 pKa = 8.03TVFDD297 pKa = 4.27EE298 pKa = 4.38EE299 pKa = 4.68SSRR302 pKa = 11.84HH303 pKa = 4.64NLRR306 pKa = 11.84EE307 pKa = 4.13KK308 pKa = 10.37IVSAFVAANEE318 pKa = 4.1DD319 pKa = 3.21FRR321 pKa = 11.84LADD324 pKa = 3.27SGARR328 pKa = 11.84QTLVNNARR336 pKa = 11.84RR337 pKa = 11.84QIKK340 pKa = 10.01LLEE343 pKa = 4.09RR344 pKa = 11.84QMPQRR349 pKa = 11.84LNLRR353 pKa = 11.84QKK355 pKa = 8.12TGPVRR360 pKa = 11.84TVPLRR365 pKa = 11.84WKK367 pKa = 9.96INRR370 pKa = 11.84RR371 pKa = 11.84TILNPLWSVTLSLLSEE387 pKa = 4.31SDD389 pKa = 3.26QLNPEE394 pKa = 4.13DD395 pKa = 4.13VEE397 pKa = 5.12LIQEE401 pKa = 4.29TAYY404 pKa = 10.68HH405 pKa = 5.8SVILEE410 pKa = 3.9SLLPKK415 pKa = 10.15KK416 pKa = 10.22KK417 pKa = 10.26KK418 pKa = 10.25KK419 pKa = 10.4NNSKK423 pKa = 8.5KK424 pKa = 8.06TAEE427 pKa = 4.25AVHH430 pKa = 5.55QVAEE434 pKa = 4.31AAPPPTPHH442 pKa = 7.18PSEE445 pKa = 3.95EE446 pKa = 4.2VTAEE450 pKa = 4.11EE451 pKa = 4.64LWGDD455 pKa = 3.87DD456 pKa = 4.43ASPDD460 pKa = 3.9SSWLQEE466 pKa = 4.27TQEE469 pKa = 4.03EE470 pKa = 5.11RR471 pKa = 11.84YY472 pKa = 10.14EE473 pKa = 4.13NQQ475 pKa = 2.85
Molecular weight: 52.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1666 |
291 |
475 |
416.5 |
46.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.122 ± 0.816 | 2.041 ± 0.435 |
4.502 ± 0.712 | 5.762 ± 0.949 |
3.902 ± 0.723 | 6.963 ± 0.508 |
2.041 ± 0.354 | 5.282 ± 0.734 |
6.543 ± 1.797 | 7.263 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.638 | 4.802 ± 0.452 |
6.783 ± 0.767 | 3.962 ± 0.442 |
6.783 ± 0.601 | 8.103 ± 1.392 |
5.522 ± 0.479 | 6.483 ± 0.629 |
1.981 ± 0.145 | 2.701 ± 0.834 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
