
Capybara microvirus Cap3_SP_481
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
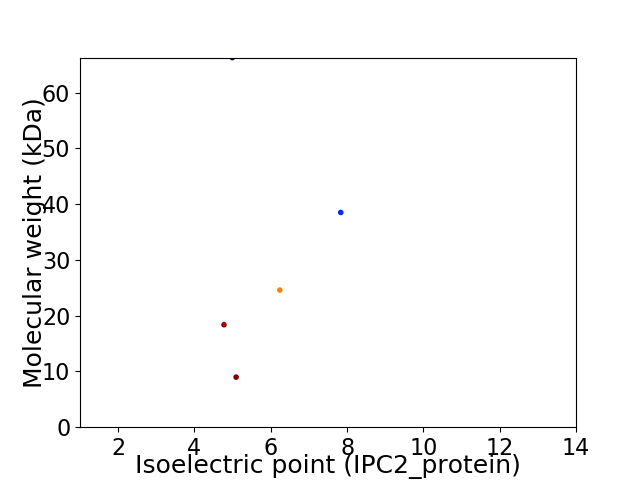
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8R9|A0A4P8W8R9_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_481 OX=2585469 PE=4 SV=1
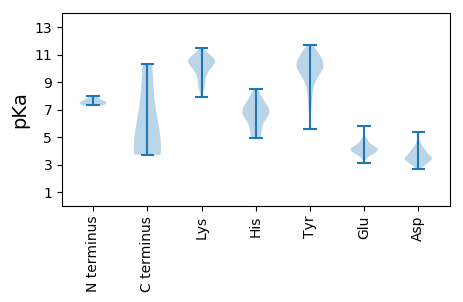
MM1 pKa = 7.55KK2 pKa = 10.28FYY4 pKa = 10.04TAYY7 pKa = 10.32NRR9 pKa = 11.84PASEE13 pKa = 3.91PTEE16 pKa = 4.36IGSSIYY22 pKa = 9.94PIYY25 pKa = 10.14EE26 pKa = 3.4PRR28 pKa = 11.84IDD30 pKa = 3.58KK31 pKa = 9.23KK32 pKa = 9.16TGRR35 pKa = 11.84KK36 pKa = 8.86ILIKK40 pKa = 10.7VGEE43 pKa = 4.02EE44 pKa = 3.58DD45 pKa = 4.02LYY47 pKa = 11.68EE48 pKa = 4.82KK49 pKa = 10.33IQEE52 pKa = 4.29SLEE55 pKa = 3.97GTKK58 pKa = 9.82IEE60 pKa = 4.42NIVRR64 pKa = 11.84RR65 pKa = 11.84VTLGDD70 pKa = 3.43PTALEE75 pKa = 3.96QRR77 pKa = 11.84IGQYY81 pKa = 11.02LDD83 pKa = 3.37LTEE86 pKa = 4.24MPEE89 pKa = 4.26NLMAAQNFILKK100 pKa = 10.53AEE102 pKa = 4.31AEE104 pKa = 4.27FEE106 pKa = 4.17KK107 pKa = 11.04LPLEE111 pKa = 3.86IRR113 pKa = 11.84KK114 pKa = 8.81EE115 pKa = 4.11FNYY118 pKa = 9.43STEE121 pKa = 4.46EE122 pKa = 3.92YY123 pKa = 9.65VSAYY127 pKa = 10.2GSEE130 pKa = 3.72EE131 pKa = 3.55WANRR135 pKa = 11.84VGLTKK140 pKa = 10.39VQEE143 pKa = 4.47EE144 pKa = 4.26IKK146 pKa = 10.49EE147 pKa = 4.01AATGVTADD155 pKa = 4.02DD156 pKa = 4.01QRR158 pKa = 11.84NEE160 pKa = 3.71
MM1 pKa = 7.55KK2 pKa = 10.28FYY4 pKa = 10.04TAYY7 pKa = 10.32NRR9 pKa = 11.84PASEE13 pKa = 3.91PTEE16 pKa = 4.36IGSSIYY22 pKa = 9.94PIYY25 pKa = 10.14EE26 pKa = 3.4PRR28 pKa = 11.84IDD30 pKa = 3.58KK31 pKa = 9.23KK32 pKa = 9.16TGRR35 pKa = 11.84KK36 pKa = 8.86ILIKK40 pKa = 10.7VGEE43 pKa = 4.02EE44 pKa = 3.58DD45 pKa = 4.02LYY47 pKa = 11.68EE48 pKa = 4.82KK49 pKa = 10.33IQEE52 pKa = 4.29SLEE55 pKa = 3.97GTKK58 pKa = 9.82IEE60 pKa = 4.42NIVRR64 pKa = 11.84RR65 pKa = 11.84VTLGDD70 pKa = 3.43PTALEE75 pKa = 3.96QRR77 pKa = 11.84IGQYY81 pKa = 11.02LDD83 pKa = 3.37LTEE86 pKa = 4.24MPEE89 pKa = 4.26NLMAAQNFILKK100 pKa = 10.53AEE102 pKa = 4.31AEE104 pKa = 4.27FEE106 pKa = 4.17KK107 pKa = 11.04LPLEE111 pKa = 3.86IRR113 pKa = 11.84KK114 pKa = 8.81EE115 pKa = 4.11FNYY118 pKa = 9.43STEE121 pKa = 4.46EE122 pKa = 3.92YY123 pKa = 9.65VSAYY127 pKa = 10.2GSEE130 pKa = 3.72EE131 pKa = 3.55WANRR135 pKa = 11.84VGLTKK140 pKa = 10.39VQEE143 pKa = 4.47EE144 pKa = 4.26IKK146 pKa = 10.49EE147 pKa = 4.01AATGVTADD155 pKa = 4.02DD156 pKa = 4.01QRR158 pKa = 11.84NEE160 pKa = 3.71
Molecular weight: 18.38 kDa
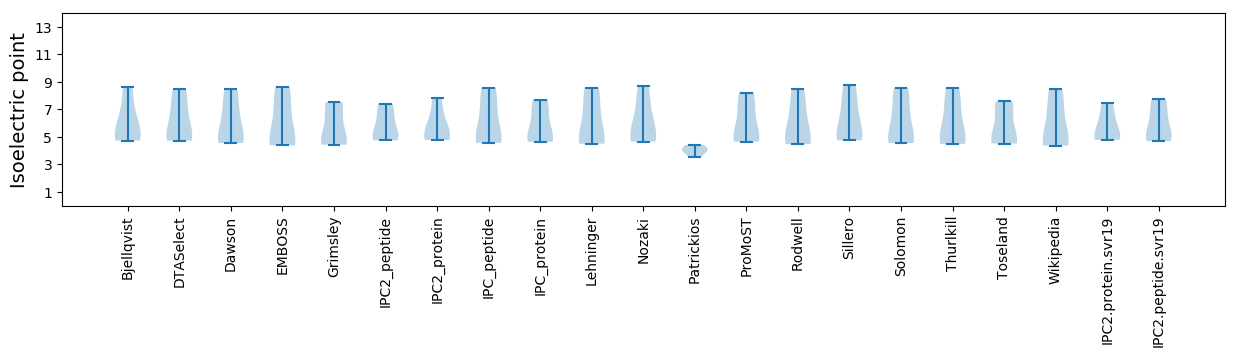
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6B1|A0A4P8W6B1_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_481 OX=2585469 PE=4 SV=1
MM1 pKa = 7.55ACSSPLTGIIDD12 pKa = 3.73QKK14 pKa = 11.47GKK16 pKa = 8.66ITFEE20 pKa = 4.2SIQLKK25 pKa = 10.46DD26 pKa = 3.36GEE28 pKa = 4.72SIKK31 pKa = 10.28EE32 pKa = 3.73RR33 pKa = 11.84KK34 pKa = 8.67QRR36 pKa = 11.84LEE38 pKa = 3.49NSGYY42 pKa = 9.02ISKK45 pKa = 10.36IIQIPCRR52 pKa = 11.84RR53 pKa = 11.84CMGCRR58 pKa = 11.84LDD60 pKa = 4.9YY61 pKa = 11.08AKK63 pKa = 10.65EE64 pKa = 3.52WANRR68 pKa = 11.84LTLEE72 pKa = 4.53TKK74 pKa = 9.91TSEE77 pKa = 3.75NNYY80 pKa = 10.09FITLTYY86 pKa = 10.55DD87 pKa = 3.42DD88 pKa = 4.41NNIPIRR94 pKa = 11.84EE95 pKa = 4.11NKK97 pKa = 10.51GEE99 pKa = 4.29FISFPLNKK107 pKa = 9.4KK108 pKa = 9.56DD109 pKa = 5.36AQDD112 pKa = 2.8FWKK115 pKa = 10.18RR116 pKa = 11.84VRR118 pKa = 11.84AKK120 pKa = 10.69YY121 pKa = 9.81PEE123 pKa = 3.76PHH125 pKa = 6.31IKK127 pKa = 10.66YY128 pKa = 9.61FMCGEE133 pKa = 4.17YY134 pKa = 10.86GEE136 pKa = 4.53EE137 pKa = 3.89TGRR140 pKa = 11.84PHH142 pKa = 5.31YY143 pKa = 10.24HH144 pKa = 6.75AIIYY148 pKa = 7.29NAPWLNDD155 pKa = 3.12LKK157 pKa = 11.04YY158 pKa = 10.92YY159 pKa = 10.54KK160 pKa = 10.73NNEE163 pKa = 3.66FGDD166 pKa = 4.03ALFHH170 pKa = 7.68SEE172 pKa = 4.82ILNKK176 pKa = 10.31LWGKK180 pKa = 11.13GDD182 pKa = 3.6TTVGDD187 pKa = 4.0VTWNSSSYY195 pKa = 7.73VARR198 pKa = 11.84YY199 pKa = 6.88ITKK202 pKa = 9.77KK203 pKa = 10.58QYY205 pKa = 9.87GDD207 pKa = 3.29KK208 pKa = 10.82AAEE211 pKa = 4.21HH212 pKa = 6.39YY213 pKa = 9.13EE214 pKa = 3.98NLGLEE219 pKa = 4.38PEE221 pKa = 4.41FVTMSLKK228 pKa = 10.72PMIGQEE234 pKa = 4.14YY235 pKa = 9.04YY236 pKa = 10.39DD237 pKa = 3.35QHH239 pKa = 8.46KK240 pKa = 9.26EE241 pKa = 3.69QIYY244 pKa = 9.98KK245 pKa = 10.3NDD247 pKa = 4.11YY248 pKa = 10.18IWINCKK254 pKa = 10.81GEE256 pKa = 4.21TIKK259 pKa = 10.64IKK261 pKa = 10.31PPRR264 pKa = 11.84IYY266 pKa = 10.4DD267 pKa = 3.7LKK269 pKa = 11.32YY270 pKa = 10.16EE271 pKa = 4.12IEE273 pKa = 4.1NPEE276 pKa = 3.72RR277 pKa = 11.84MKK279 pKa = 10.5EE280 pKa = 3.81IKK282 pKa = 9.89LQRR285 pKa = 11.84QLRR288 pKa = 11.84QEE290 pKa = 3.84AATAEE295 pKa = 4.43IINNTGYY302 pKa = 10.3IDD304 pKa = 3.98GAQMQRR310 pKa = 11.84EE311 pKa = 4.36IKK313 pKa = 10.49ARR315 pKa = 11.84ALDD318 pKa = 3.59ARR320 pKa = 11.84TANLKK325 pKa = 8.91RR326 pKa = 11.84TII328 pKa = 3.82
MM1 pKa = 7.55ACSSPLTGIIDD12 pKa = 3.73QKK14 pKa = 11.47GKK16 pKa = 8.66ITFEE20 pKa = 4.2SIQLKK25 pKa = 10.46DD26 pKa = 3.36GEE28 pKa = 4.72SIKK31 pKa = 10.28EE32 pKa = 3.73RR33 pKa = 11.84KK34 pKa = 8.67QRR36 pKa = 11.84LEE38 pKa = 3.49NSGYY42 pKa = 9.02ISKK45 pKa = 10.36IIQIPCRR52 pKa = 11.84RR53 pKa = 11.84CMGCRR58 pKa = 11.84LDD60 pKa = 4.9YY61 pKa = 11.08AKK63 pKa = 10.65EE64 pKa = 3.52WANRR68 pKa = 11.84LTLEE72 pKa = 4.53TKK74 pKa = 9.91TSEE77 pKa = 3.75NNYY80 pKa = 10.09FITLTYY86 pKa = 10.55DD87 pKa = 3.42DD88 pKa = 4.41NNIPIRR94 pKa = 11.84EE95 pKa = 4.11NKK97 pKa = 10.51GEE99 pKa = 4.29FISFPLNKK107 pKa = 9.4KK108 pKa = 9.56DD109 pKa = 5.36AQDD112 pKa = 2.8FWKK115 pKa = 10.18RR116 pKa = 11.84VRR118 pKa = 11.84AKK120 pKa = 10.69YY121 pKa = 9.81PEE123 pKa = 3.76PHH125 pKa = 6.31IKK127 pKa = 10.66YY128 pKa = 9.61FMCGEE133 pKa = 4.17YY134 pKa = 10.86GEE136 pKa = 4.53EE137 pKa = 3.89TGRR140 pKa = 11.84PHH142 pKa = 5.31YY143 pKa = 10.24HH144 pKa = 6.75AIIYY148 pKa = 7.29NAPWLNDD155 pKa = 3.12LKK157 pKa = 11.04YY158 pKa = 10.92YY159 pKa = 10.54KK160 pKa = 10.73NNEE163 pKa = 3.66FGDD166 pKa = 4.03ALFHH170 pKa = 7.68SEE172 pKa = 4.82ILNKK176 pKa = 10.31LWGKK180 pKa = 11.13GDD182 pKa = 3.6TTVGDD187 pKa = 4.0VTWNSSSYY195 pKa = 7.73VARR198 pKa = 11.84YY199 pKa = 6.88ITKK202 pKa = 9.77KK203 pKa = 10.58QYY205 pKa = 9.87GDD207 pKa = 3.29KK208 pKa = 10.82AAEE211 pKa = 4.21HH212 pKa = 6.39YY213 pKa = 9.13EE214 pKa = 3.98NLGLEE219 pKa = 4.38PEE221 pKa = 4.41FVTMSLKK228 pKa = 10.72PMIGQEE234 pKa = 4.14YY235 pKa = 9.04YY236 pKa = 10.39DD237 pKa = 3.35QHH239 pKa = 8.46KK240 pKa = 9.26EE241 pKa = 3.69QIYY244 pKa = 9.98KK245 pKa = 10.3NDD247 pKa = 4.11YY248 pKa = 10.18IWINCKK254 pKa = 10.81GEE256 pKa = 4.21TIKK259 pKa = 10.64IKK261 pKa = 10.31PPRR264 pKa = 11.84IYY266 pKa = 10.4DD267 pKa = 3.7LKK269 pKa = 11.32YY270 pKa = 10.16EE271 pKa = 4.12IEE273 pKa = 4.1NPEE276 pKa = 3.72RR277 pKa = 11.84MKK279 pKa = 10.5EE280 pKa = 3.81IKK282 pKa = 9.89LQRR285 pKa = 11.84QLRR288 pKa = 11.84QEE290 pKa = 3.84AATAEE295 pKa = 4.43IINNTGYY302 pKa = 10.3IDD304 pKa = 3.98GAQMQRR310 pKa = 11.84EE311 pKa = 4.36IKK313 pKa = 10.49ARR315 pKa = 11.84ALDD318 pKa = 3.59ARR320 pKa = 11.84TANLKK325 pKa = 8.91RR326 pKa = 11.84TII328 pKa = 3.82
Molecular weight: 38.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1379 |
78 |
580 |
275.8 |
31.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.759 ± 1.103 | 0.725 ± 0.366 |
4.496 ± 0.34 | 8.484 ± 1.682 |
3.481 ± 1.044 | 7.179 ± 0.916 |
1.378 ± 0.385 | 7.179 ± 1.261 |
6.599 ± 1.253 | 5.729 ± 0.418 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.191 ± 0.471 | 6.526 ± 1.147 |
4.206 ± 0.923 | 4.641 ± 0.376 |
4.206 ± 0.71 | 6.164 ± 1.601 |
7.179 ± 0.653 | 3.191 ± 0.649 |
2.03 ± 0.412 | 5.656 ± 0.455 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
