
Capybara microvirus Cap3_SP_386
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
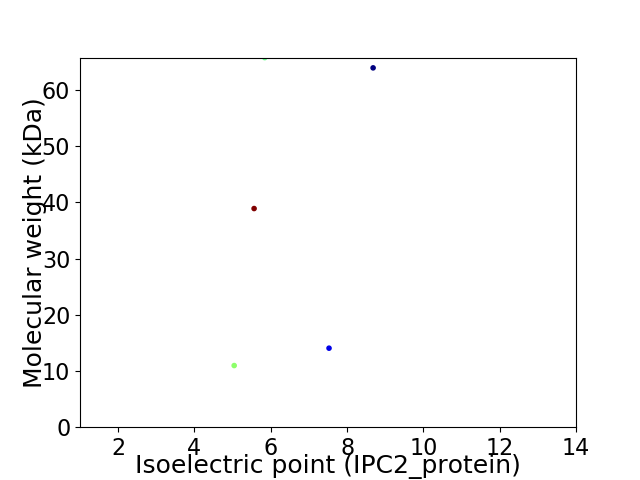
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
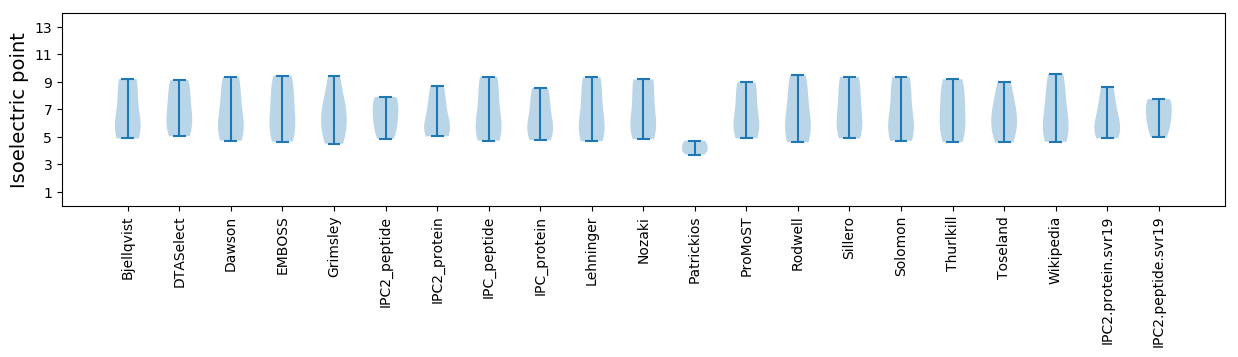
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVS7|A0A4V1FVS7_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_386 OX=2585440 PE=4 SV=1
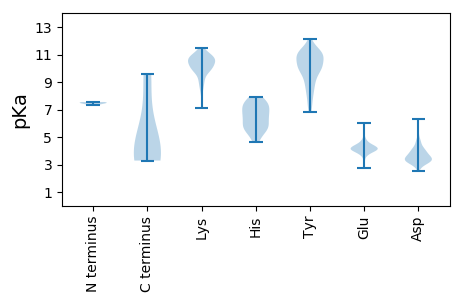
MM1 pKa = 7.34YY2 pKa = 10.77NFIKK6 pKa = 10.29KK7 pKa = 10.03RR8 pKa = 11.84PLEE11 pKa = 4.09QYY13 pKa = 8.14EE14 pKa = 4.6SPVITIQDD22 pKa = 4.05FSTVPDD28 pKa = 3.55EE29 pKa = 4.75NGNKK33 pKa = 9.3VAVFSPSTPKK43 pKa = 9.5IDD45 pKa = 3.48KK46 pKa = 10.83DD47 pKa = 3.16SDD49 pKa = 3.48YY50 pKa = 11.72RR51 pKa = 11.84LMLVGNLIKK60 pKa = 10.8SGINPVSGIFVNPLNRR76 pKa = 11.84LSVDD80 pKa = 4.65SIAQDD85 pKa = 3.88YY86 pKa = 7.9ITSLNQSINEE96 pKa = 4.19TLL98 pKa = 3.29
MM1 pKa = 7.34YY2 pKa = 10.77NFIKK6 pKa = 10.29KK7 pKa = 10.03RR8 pKa = 11.84PLEE11 pKa = 4.09QYY13 pKa = 8.14EE14 pKa = 4.6SPVITIQDD22 pKa = 4.05FSTVPDD28 pKa = 3.55EE29 pKa = 4.75NGNKK33 pKa = 9.3VAVFSPSTPKK43 pKa = 9.5IDD45 pKa = 3.48KK46 pKa = 10.83DD47 pKa = 3.16SDD49 pKa = 3.48YY50 pKa = 11.72RR51 pKa = 11.84LMLVGNLIKK60 pKa = 10.8SGINPVSGIFVNPLNRR76 pKa = 11.84LSVDD80 pKa = 4.65SIAQDD85 pKa = 3.88YY86 pKa = 7.9ITSLNQSINEE96 pKa = 4.19TLL98 pKa = 3.29
Molecular weight: 10.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W599|A0A4P8W599_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_386 OX=2585440 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.1EE3 pKa = 3.99VITIPEE9 pKa = 4.11VTDD12 pKa = 3.87FSYY15 pKa = 11.02RR16 pKa = 11.84FNRR19 pKa = 11.84EE20 pKa = 2.76KK21 pKa = 11.19DD22 pKa = 3.2FLIRR26 pKa = 11.84LNYY29 pKa = 10.12EE30 pKa = 3.62YY31 pKa = 10.82LDD33 pKa = 4.71CIKK36 pKa = 10.78QGGKK40 pKa = 9.98CLFYY44 pKa = 10.7TLTYY48 pKa = 10.0TNQALPKK55 pKa = 8.29IYY57 pKa = 9.86GVPVHH62 pKa = 7.47DD63 pKa = 4.0YY64 pKa = 10.01HH65 pKa = 7.92DD66 pKa = 3.96LRR68 pKa = 11.84SFVNGSFYY76 pKa = 11.01KK77 pKa = 10.34QVSKK81 pKa = 9.67TCNLRR86 pKa = 11.84YY87 pKa = 9.59FISCEE92 pKa = 3.91SGEE95 pKa = 4.5GKK97 pKa = 10.37GKK99 pKa = 10.53RR100 pKa = 11.84GFANNSHH107 pKa = 4.67YY108 pKa = 10.76HH109 pKa = 5.57ILFFLFPKK117 pKa = 9.93EE118 pKa = 4.32GKK120 pKa = 9.89SLLQPYY126 pKa = 7.46EE127 pKa = 3.57FRR129 pKa = 11.84HH130 pKa = 4.86YY131 pKa = 9.92VRR133 pKa = 11.84KK134 pKa = 9.19YY135 pKa = 7.54WQGFDD140 pKa = 3.43EE141 pKa = 4.96SVCFEE146 pKa = 4.4DD147 pKa = 6.33FRR149 pKa = 11.84NCKK152 pKa = 10.11YY153 pKa = 10.92GIARR157 pKa = 11.84EE158 pKa = 3.91GDD160 pKa = 3.31NLGEE164 pKa = 4.17VTSINALQYY173 pKa = 8.59VVKK176 pKa = 10.82YY177 pKa = 6.97VTKK180 pKa = 10.45DD181 pKa = 3.19LNSISNEE188 pKa = 3.33KK189 pKa = 10.53KK190 pKa = 9.87IFDD193 pKa = 3.87RR194 pKa = 11.84YY195 pKa = 10.05EE196 pKa = 3.99SYY198 pKa = 11.26LNKK201 pKa = 9.83ISYY204 pKa = 9.7LPKK207 pKa = 10.29SYY209 pKa = 11.05SDD211 pKa = 3.8YY212 pKa = 11.43VDD214 pKa = 3.23TFFNGAVLVPNFDD227 pKa = 3.1FATRR231 pKa = 11.84LFRR234 pKa = 11.84RR235 pKa = 11.84YY236 pKa = 9.18IHH238 pKa = 5.67TLKK241 pKa = 10.69SKK243 pKa = 10.32YY244 pKa = 7.24NTRR247 pKa = 11.84LVSRR251 pKa = 11.84SNLLLLINEE260 pKa = 4.4EE261 pKa = 4.28YY262 pKa = 11.02KK263 pKa = 11.16NFLISTKK270 pKa = 10.59KK271 pKa = 10.35KK272 pKa = 8.62ICFDD276 pKa = 3.33EE277 pKa = 5.25FIIRR281 pKa = 11.84EE282 pKa = 4.0ASLLTFFEE290 pKa = 3.93QWRR293 pKa = 11.84RR294 pKa = 11.84QQVSEE299 pKa = 4.24KK300 pKa = 10.12LQEE303 pKa = 3.92FKK305 pKa = 10.95KK306 pKa = 10.42IFRR309 pKa = 11.84NRR311 pKa = 11.84YY312 pKa = 6.8SSKK315 pKa = 9.97PRR317 pKa = 11.84FSHH320 pKa = 6.17GLGKK324 pKa = 10.22IGLDD328 pKa = 3.67TVDD331 pKa = 2.98KK332 pKa = 10.4SYY334 pKa = 10.92PIFVDD339 pKa = 3.46PCTNKK344 pKa = 9.74PVRR347 pKa = 11.84IPLYY351 pKa = 9.05YY352 pKa = 10.11FRR354 pKa = 11.84HH355 pKa = 6.52LFTDD359 pKa = 3.88VKK361 pKa = 10.57KK362 pKa = 11.1LNGNNVYY369 pKa = 10.45VLNEE373 pKa = 4.1DD374 pKa = 3.66GIKK377 pKa = 10.58YY378 pKa = 10.11KK379 pKa = 10.49EE380 pKa = 4.23SKK382 pKa = 10.65LSEE385 pKa = 4.16KK386 pKa = 10.71LNYY389 pKa = 10.34LADD392 pKa = 3.69QLIKK396 pKa = 10.73SFYY399 pKa = 10.06AASKK403 pKa = 10.36LPSDD407 pKa = 3.43IQLLNFGRR415 pKa = 11.84YY416 pKa = 8.27IDD418 pKa = 3.36IAFPNRR424 pKa = 11.84KK425 pKa = 9.12YY426 pKa = 10.98VDD428 pKa = 3.38DD429 pKa = 3.8YY430 pKa = 12.12ARR432 pKa = 11.84FKK434 pKa = 11.0LIYY437 pKa = 9.1EE438 pKa = 4.16GRR440 pKa = 11.84KK441 pKa = 9.35FPISEE446 pKa = 4.22FNNPNLLDD454 pKa = 4.49DD455 pKa = 4.15SLTYY459 pKa = 10.33RR460 pKa = 11.84SNLFCSAWYY469 pKa = 9.04FPPGLVTDD477 pKa = 4.11YY478 pKa = 11.34NPDD481 pKa = 3.56YY482 pKa = 11.2LEE484 pKa = 4.33YY485 pKa = 10.33AYY487 pKa = 10.41HH488 pKa = 7.56PYY490 pKa = 10.24FLQNIEE496 pKa = 4.11LFSYY500 pKa = 10.52FDD502 pKa = 4.78RR503 pKa = 11.84ILSYY507 pKa = 10.25FANNKK512 pKa = 8.84NVVYY516 pKa = 8.52WQKK519 pKa = 11.1YY520 pKa = 8.17NDD522 pKa = 3.32WKK524 pKa = 9.69KK525 pKa = 8.29TKK527 pKa = 10.58NYY529 pKa = 9.78INSLSII535 pKa = 3.67
MM1 pKa = 7.54KK2 pKa = 10.1EE3 pKa = 3.99VITIPEE9 pKa = 4.11VTDD12 pKa = 3.87FSYY15 pKa = 11.02RR16 pKa = 11.84FNRR19 pKa = 11.84EE20 pKa = 2.76KK21 pKa = 11.19DD22 pKa = 3.2FLIRR26 pKa = 11.84LNYY29 pKa = 10.12EE30 pKa = 3.62YY31 pKa = 10.82LDD33 pKa = 4.71CIKK36 pKa = 10.78QGGKK40 pKa = 9.98CLFYY44 pKa = 10.7TLTYY48 pKa = 10.0TNQALPKK55 pKa = 8.29IYY57 pKa = 9.86GVPVHH62 pKa = 7.47DD63 pKa = 4.0YY64 pKa = 10.01HH65 pKa = 7.92DD66 pKa = 3.96LRR68 pKa = 11.84SFVNGSFYY76 pKa = 11.01KK77 pKa = 10.34QVSKK81 pKa = 9.67TCNLRR86 pKa = 11.84YY87 pKa = 9.59FISCEE92 pKa = 3.91SGEE95 pKa = 4.5GKK97 pKa = 10.37GKK99 pKa = 10.53RR100 pKa = 11.84GFANNSHH107 pKa = 4.67YY108 pKa = 10.76HH109 pKa = 5.57ILFFLFPKK117 pKa = 9.93EE118 pKa = 4.32GKK120 pKa = 9.89SLLQPYY126 pKa = 7.46EE127 pKa = 3.57FRR129 pKa = 11.84HH130 pKa = 4.86YY131 pKa = 9.92VRR133 pKa = 11.84KK134 pKa = 9.19YY135 pKa = 7.54WQGFDD140 pKa = 3.43EE141 pKa = 4.96SVCFEE146 pKa = 4.4DD147 pKa = 6.33FRR149 pKa = 11.84NCKK152 pKa = 10.11YY153 pKa = 10.92GIARR157 pKa = 11.84EE158 pKa = 3.91GDD160 pKa = 3.31NLGEE164 pKa = 4.17VTSINALQYY173 pKa = 8.59VVKK176 pKa = 10.82YY177 pKa = 6.97VTKK180 pKa = 10.45DD181 pKa = 3.19LNSISNEE188 pKa = 3.33KK189 pKa = 10.53KK190 pKa = 9.87IFDD193 pKa = 3.87RR194 pKa = 11.84YY195 pKa = 10.05EE196 pKa = 3.99SYY198 pKa = 11.26LNKK201 pKa = 9.83ISYY204 pKa = 9.7LPKK207 pKa = 10.29SYY209 pKa = 11.05SDD211 pKa = 3.8YY212 pKa = 11.43VDD214 pKa = 3.23TFFNGAVLVPNFDD227 pKa = 3.1FATRR231 pKa = 11.84LFRR234 pKa = 11.84RR235 pKa = 11.84YY236 pKa = 9.18IHH238 pKa = 5.67TLKK241 pKa = 10.69SKK243 pKa = 10.32YY244 pKa = 7.24NTRR247 pKa = 11.84LVSRR251 pKa = 11.84SNLLLLINEE260 pKa = 4.4EE261 pKa = 4.28YY262 pKa = 11.02KK263 pKa = 11.16NFLISTKK270 pKa = 10.59KK271 pKa = 10.35KK272 pKa = 8.62ICFDD276 pKa = 3.33EE277 pKa = 5.25FIIRR281 pKa = 11.84EE282 pKa = 4.0ASLLTFFEE290 pKa = 3.93QWRR293 pKa = 11.84RR294 pKa = 11.84QQVSEE299 pKa = 4.24KK300 pKa = 10.12LQEE303 pKa = 3.92FKK305 pKa = 10.95KK306 pKa = 10.42IFRR309 pKa = 11.84NRR311 pKa = 11.84YY312 pKa = 6.8SSKK315 pKa = 9.97PRR317 pKa = 11.84FSHH320 pKa = 6.17GLGKK324 pKa = 10.22IGLDD328 pKa = 3.67TVDD331 pKa = 2.98KK332 pKa = 10.4SYY334 pKa = 10.92PIFVDD339 pKa = 3.46PCTNKK344 pKa = 9.74PVRR347 pKa = 11.84IPLYY351 pKa = 9.05YY352 pKa = 10.11FRR354 pKa = 11.84HH355 pKa = 6.52LFTDD359 pKa = 3.88VKK361 pKa = 10.57KK362 pKa = 11.1LNGNNVYY369 pKa = 10.45VLNEE373 pKa = 4.1DD374 pKa = 3.66GIKK377 pKa = 10.58YY378 pKa = 10.11KK379 pKa = 10.49EE380 pKa = 4.23SKK382 pKa = 10.65LSEE385 pKa = 4.16KK386 pKa = 10.71LNYY389 pKa = 10.34LADD392 pKa = 3.69QLIKK396 pKa = 10.73SFYY399 pKa = 10.06AASKK403 pKa = 10.36LPSDD407 pKa = 3.43IQLLNFGRR415 pKa = 11.84YY416 pKa = 8.27IDD418 pKa = 3.36IAFPNRR424 pKa = 11.84KK425 pKa = 9.12YY426 pKa = 10.98VDD428 pKa = 3.38DD429 pKa = 3.8YY430 pKa = 12.12ARR432 pKa = 11.84FKK434 pKa = 11.0LIYY437 pKa = 9.1EE438 pKa = 4.16GRR440 pKa = 11.84KK441 pKa = 9.35FPISEE446 pKa = 4.22FNNPNLLDD454 pKa = 4.49DD455 pKa = 4.15SLTYY459 pKa = 10.33RR460 pKa = 11.84SNLFCSAWYY469 pKa = 9.04FPPGLVTDD477 pKa = 4.11YY478 pKa = 11.34NPDD481 pKa = 3.56YY482 pKa = 11.2LEE484 pKa = 4.33YY485 pKa = 10.33AYY487 pKa = 10.41HH488 pKa = 7.56PYY490 pKa = 10.24FLQNIEE496 pKa = 4.11LFSYY500 pKa = 10.52FDD502 pKa = 4.78RR503 pKa = 11.84ILSYY507 pKa = 10.25FANNKK512 pKa = 8.84NVVYY516 pKa = 8.52WQKK519 pKa = 11.1YY520 pKa = 8.17NDD522 pKa = 3.32WKK524 pKa = 9.69KK525 pKa = 8.29TKK527 pKa = 10.58NYY529 pKa = 9.78INSLSII535 pKa = 3.67
Molecular weight: 63.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1686 |
98 |
580 |
337.2 |
38.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.567 ± 1.163 | 0.949 ± 0.331 |
5.516 ± 0.417 | 5.219 ± 0.633 |
6.168 ± 1.102 | 4.686 ± 0.904 |
1.839 ± 0.378 | 6.999 ± 1.148 |
6.821 ± 0.995 | 9.075 ± 0.584 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.246 ± 0.411 | 7.711 ± 0.986 |
3.321 ± 0.464 | 4.152 ± 1.06 |
4.508 ± 0.562 | 10.202 ± 1.165 |
5.279 ± 0.57 | 5.219 ± 0.512 |
0.712 ± 0.192 | 5.813 ± 1.591 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
