
Blastococcus saxobsidens (strain DD2)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; Blastococcus saxobsidens
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
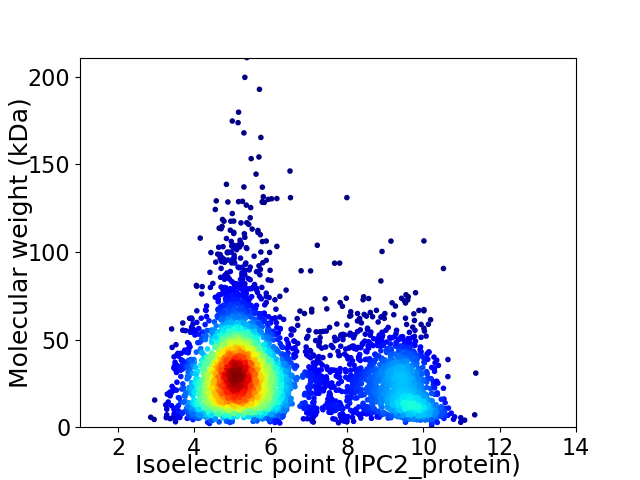
Virtual 2D-PAGE plot for 4791 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
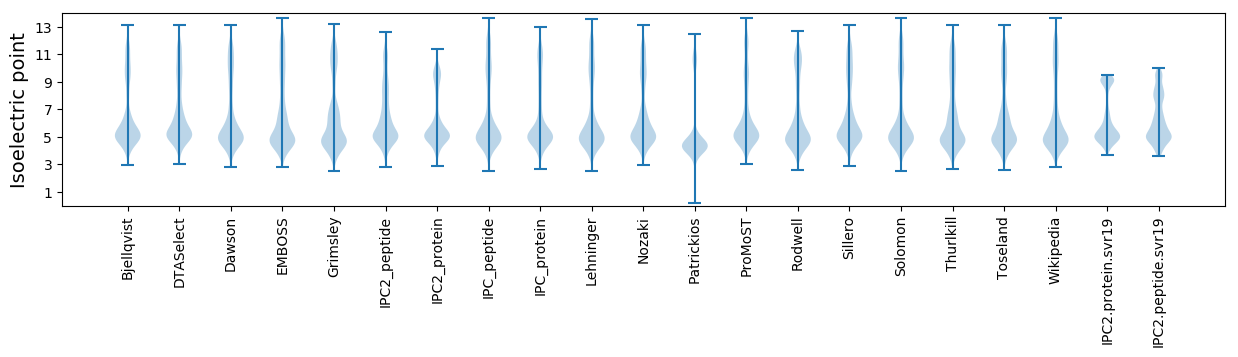
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H6RUH3|H6RUH3_BLASD ESAT-6-like protein OS=Blastococcus saxobsidens (strain DD2) OX=1146883 GN=BLASA_0990 PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.3LSKK5 pKa = 10.55RR6 pKa = 11.84NSGLVATAIAAALLMTACGGGDD28 pKa = 3.62DD29 pKa = 4.4EE30 pKa = 5.63GGEE33 pKa = 4.61GGGSADD39 pKa = 3.43GGGSYY44 pKa = 10.84SLAINNPEE52 pKa = 3.88NPLVPGNTTEE62 pKa = 4.37SEE64 pKa = 4.24GAQVIKK70 pKa = 10.71ALWTGLIEE78 pKa = 4.05YY79 pKa = 10.58AEE81 pKa = 4.8DD82 pKa = 4.83GEE84 pKa = 4.44VQYY87 pKa = 11.03TGIAEE92 pKa = 4.42SIEE95 pKa = 4.35SDD97 pKa = 5.33DD98 pKa = 3.61NTTWTITLKK107 pKa = 10.95DD108 pKa = 3.06GWTFHH113 pKa = 7.56DD114 pKa = 4.47GTPVTASSFVDD125 pKa = 3.19AWNYY129 pKa = 7.22TAYY132 pKa = 10.7SPNAQGGSYY141 pKa = 10.2FLSNIVGYY149 pKa = 10.65KK150 pKa = 10.18DD151 pKa = 3.5LQAPTDD157 pKa = 3.79DD158 pKa = 4.21AGEE161 pKa = 4.11ITGDD165 pKa = 3.63PVATEE170 pKa = 3.92MTGLEE175 pKa = 4.29VVDD178 pKa = 4.51DD179 pKa = 3.75QTFTVTLNDD188 pKa = 3.84PFAQFPVTVGYY199 pKa = 10.29NAFFPMAEE207 pKa = 4.37AFFEE211 pKa = 4.57DD212 pKa = 4.58PEE214 pKa = 4.68AAALDD219 pKa = 4.2TPIGTGPFQAEE230 pKa = 4.72GPFEE234 pKa = 4.36EE235 pKa = 5.25GVGITLSKK243 pKa = 10.31YY244 pKa = 10.65DD245 pKa = 4.37DD246 pKa = 3.7YY247 pKa = 12.02AGEE250 pKa = 4.16NEE252 pKa = 4.39ANAEE256 pKa = 4.05TLEE259 pKa = 4.43IDD261 pKa = 3.95VYY263 pKa = 11.28SEE265 pKa = 3.47ITTSYY270 pKa = 11.17RR271 pKa = 11.84DD272 pKa = 3.65VQGGNLDD279 pKa = 3.72VVKK282 pKa = 10.44DD283 pKa = 3.84IPADD287 pKa = 3.87VIATAPAEE295 pKa = 4.21FGDD298 pKa = 3.98RR299 pKa = 11.84FIEE302 pKa = 4.46RR303 pKa = 11.84PSSSFTYY310 pKa = 10.4LGVPTYY316 pKa = 10.77DD317 pKa = 3.36PRR319 pKa = 11.84FADD322 pKa = 3.13KK323 pKa = 10.42RR324 pKa = 11.84VRR326 pKa = 11.84QAFSMAIDD334 pKa = 3.82RR335 pKa = 11.84GAITDD340 pKa = 4.72AIFNGTFTPATDD352 pKa = 5.04AIAPVVDD359 pKa = 5.17GYY361 pKa = 11.49RR362 pKa = 11.84EE363 pKa = 4.25GACEE367 pKa = 3.86YY368 pKa = 11.03CEE370 pKa = 5.23LDD372 pKa = 3.89VEE374 pKa = 5.0RR375 pKa = 11.84ANQLLDD381 pKa = 3.26EE382 pKa = 5.15AGFDD386 pKa = 3.15RR387 pKa = 11.84SMPVDD392 pKa = 3.49LWFNAGASHH401 pKa = 6.94DD402 pKa = 3.73AWVEE406 pKa = 3.74AVGNQLRR413 pKa = 11.84EE414 pKa = 4.05NLGIEE419 pKa = 4.1YY420 pKa = 9.01TLQGNLEE427 pKa = 4.01FAEE430 pKa = 4.24YY431 pKa = 10.78LPLLDD436 pKa = 4.45SKK438 pKa = 11.4GITGPFRR445 pKa = 11.84LGWAMDD451 pKa = 3.73YY452 pKa = 10.19PSPQNYY458 pKa = 9.97LEE460 pKa = 4.44PLYY463 pKa = 10.24STAALPPAGSNVTFYY478 pKa = 11.46SNPEE482 pKa = 3.64FDD484 pKa = 4.25QLIAQGNQAEE494 pKa = 4.57TNEE497 pKa = 4.11EE498 pKa = 4.21AIDD501 pKa = 4.33FYY503 pKa = 11.52QQADD507 pKa = 4.22DD508 pKa = 5.75IILEE512 pKa = 4.37DD513 pKa = 3.78MPIIPMFFGVEE524 pKa = 4.07QIVHH528 pKa = 5.93SEE530 pKa = 4.08DD531 pKa = 3.39VSDD534 pKa = 4.04VVIDD538 pKa = 3.42VFGQIDD544 pKa = 3.65TARR547 pKa = 11.84VKK549 pKa = 11.06VNN551 pKa = 3.1
MM1 pKa = 7.5KK2 pKa = 10.3LSKK5 pKa = 10.55RR6 pKa = 11.84NSGLVATAIAAALLMTACGGGDD28 pKa = 3.62DD29 pKa = 4.4EE30 pKa = 5.63GGEE33 pKa = 4.61GGGSADD39 pKa = 3.43GGGSYY44 pKa = 10.84SLAINNPEE52 pKa = 3.88NPLVPGNTTEE62 pKa = 4.37SEE64 pKa = 4.24GAQVIKK70 pKa = 10.71ALWTGLIEE78 pKa = 4.05YY79 pKa = 10.58AEE81 pKa = 4.8DD82 pKa = 4.83GEE84 pKa = 4.44VQYY87 pKa = 11.03TGIAEE92 pKa = 4.42SIEE95 pKa = 4.35SDD97 pKa = 5.33DD98 pKa = 3.61NTTWTITLKK107 pKa = 10.95DD108 pKa = 3.06GWTFHH113 pKa = 7.56DD114 pKa = 4.47GTPVTASSFVDD125 pKa = 3.19AWNYY129 pKa = 7.22TAYY132 pKa = 10.7SPNAQGGSYY141 pKa = 10.2FLSNIVGYY149 pKa = 10.65KK150 pKa = 10.18DD151 pKa = 3.5LQAPTDD157 pKa = 3.79DD158 pKa = 4.21AGEE161 pKa = 4.11ITGDD165 pKa = 3.63PVATEE170 pKa = 3.92MTGLEE175 pKa = 4.29VVDD178 pKa = 4.51DD179 pKa = 3.75QTFTVTLNDD188 pKa = 3.84PFAQFPVTVGYY199 pKa = 10.29NAFFPMAEE207 pKa = 4.37AFFEE211 pKa = 4.57DD212 pKa = 4.58PEE214 pKa = 4.68AAALDD219 pKa = 4.2TPIGTGPFQAEE230 pKa = 4.72GPFEE234 pKa = 4.36EE235 pKa = 5.25GVGITLSKK243 pKa = 10.31YY244 pKa = 10.65DD245 pKa = 4.37DD246 pKa = 3.7YY247 pKa = 12.02AGEE250 pKa = 4.16NEE252 pKa = 4.39ANAEE256 pKa = 4.05TLEE259 pKa = 4.43IDD261 pKa = 3.95VYY263 pKa = 11.28SEE265 pKa = 3.47ITTSYY270 pKa = 11.17RR271 pKa = 11.84DD272 pKa = 3.65VQGGNLDD279 pKa = 3.72VVKK282 pKa = 10.44DD283 pKa = 3.84IPADD287 pKa = 3.87VIATAPAEE295 pKa = 4.21FGDD298 pKa = 3.98RR299 pKa = 11.84FIEE302 pKa = 4.46RR303 pKa = 11.84PSSSFTYY310 pKa = 10.4LGVPTYY316 pKa = 10.77DD317 pKa = 3.36PRR319 pKa = 11.84FADD322 pKa = 3.13KK323 pKa = 10.42RR324 pKa = 11.84VRR326 pKa = 11.84QAFSMAIDD334 pKa = 3.82RR335 pKa = 11.84GAITDD340 pKa = 4.72AIFNGTFTPATDD352 pKa = 5.04AIAPVVDD359 pKa = 5.17GYY361 pKa = 11.49RR362 pKa = 11.84EE363 pKa = 4.25GACEE367 pKa = 3.86YY368 pKa = 11.03CEE370 pKa = 5.23LDD372 pKa = 3.89VEE374 pKa = 5.0RR375 pKa = 11.84ANQLLDD381 pKa = 3.26EE382 pKa = 5.15AGFDD386 pKa = 3.15RR387 pKa = 11.84SMPVDD392 pKa = 3.49LWFNAGASHH401 pKa = 6.94DD402 pKa = 3.73AWVEE406 pKa = 3.74AVGNQLRR413 pKa = 11.84EE414 pKa = 4.05NLGIEE419 pKa = 4.1YY420 pKa = 9.01TLQGNLEE427 pKa = 4.01FAEE430 pKa = 4.24YY431 pKa = 10.78LPLLDD436 pKa = 4.45SKK438 pKa = 11.4GITGPFRR445 pKa = 11.84LGWAMDD451 pKa = 3.73YY452 pKa = 10.19PSPQNYY458 pKa = 9.97LEE460 pKa = 4.44PLYY463 pKa = 10.24STAALPPAGSNVTFYY478 pKa = 11.46SNPEE482 pKa = 3.64FDD484 pKa = 4.25QLIAQGNQAEE494 pKa = 4.57TNEE497 pKa = 4.11EE498 pKa = 4.21AIDD501 pKa = 4.33FYY503 pKa = 11.52QQADD507 pKa = 4.22DD508 pKa = 5.75IILEE512 pKa = 4.37DD513 pKa = 3.78MPIIPMFFGVEE524 pKa = 4.07QIVHH528 pKa = 5.93SEE530 pKa = 4.08DD531 pKa = 3.39VSDD534 pKa = 4.04VVIDD538 pKa = 3.42VFGQIDD544 pKa = 3.65TARR547 pKa = 11.84VKK549 pKa = 11.06VNN551 pKa = 3.1
Molecular weight: 59.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6RPY1|H6RPY1_BLASD Lysine--tRNA ligase OS=Blastococcus saxobsidens (strain DD2) OX=1146883 GN=lysS PE=3 SV=1
MM1 pKa = 7.46GKK3 pKa = 9.97GGRR6 pKa = 11.84MSILGKK12 pKa = 10.69LLGTAGGAARR22 pKa = 11.84GAARR26 pKa = 11.84GGRR29 pKa = 11.84ATGRR33 pKa = 11.84PGRR36 pKa = 11.84GVPRR40 pKa = 11.84SAMGRR45 pKa = 11.84TTGRR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84GTPASSGGIAGMLGSLLRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 4.1
MM1 pKa = 7.46GKK3 pKa = 9.97GGRR6 pKa = 11.84MSILGKK12 pKa = 10.69LLGTAGGAARR22 pKa = 11.84GAARR26 pKa = 11.84GGRR29 pKa = 11.84ATGRR33 pKa = 11.84PGRR36 pKa = 11.84GVPRR40 pKa = 11.84SAMGRR45 pKa = 11.84TTGRR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84GTPASSGGIAGMLGSLLRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 4.1
Molecular weight: 7.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1451551 |
10 |
1972 |
303.0 |
32.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.145 ± 0.059 | 0.718 ± 0.009 |
6.206 ± 0.03 | 5.66 ± 0.03 |
2.569 ± 0.024 | 9.589 ± 0.03 |
2.041 ± 0.017 | 3.006 ± 0.027 |
1.409 ± 0.021 | 10.634 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.015 | 1.457 ± 0.017 |
6.242 ± 0.032 | 2.788 ± 0.019 |
8.308 ± 0.044 | 4.913 ± 0.025 |
5.916 ± 0.026 | 9.51 ± 0.031 |
1.452 ± 0.015 | 1.74 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
