
Human papillomavirus 157
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 12
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
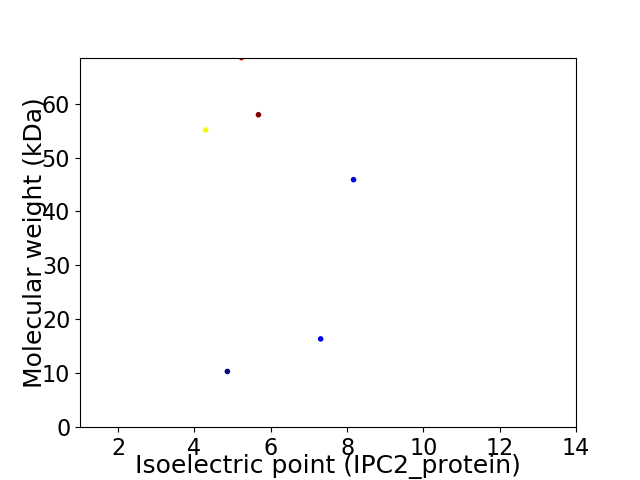
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166FK68|A0A166FK68_9PAPI Major capsid protein L1 OS=Human papillomavirus 157 OX=2259331 GN=L1 PE=3 SV=1
MM1 pKa = 7.07EE2 pKa = 5.43AKK4 pKa = 9.35TRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.5RR9 pKa = 11.84AAPDD13 pKa = 2.91QLYY16 pKa = 9.56RR17 pKa = 11.84HH18 pKa = 6.65CLQGGDD24 pKa = 5.15CIPDD28 pKa = 3.33VQNKK32 pKa = 9.5YY33 pKa = 7.08EE34 pKa = 4.1QKK36 pKa = 8.24TWADD40 pKa = 3.39VLLKK44 pKa = 10.51IFGSLLYY51 pKa = 10.37FGNLGIGTGRR61 pKa = 11.84GSGGSLGYY69 pKa = 10.22RR70 pKa = 11.84PLGTTRR76 pKa = 11.84VGEE79 pKa = 4.19VAPITPARR87 pKa = 11.84PSILIDD93 pKa = 3.34AVGPTDD99 pKa = 4.03VVPIDD104 pKa = 3.76ASTPAIVPLAEE115 pKa = 4.29GTLDD119 pKa = 3.39TGFIAPDD126 pKa = 3.64AGPGVGVEE134 pKa = 4.06EE135 pKa = 4.93LEE137 pKa = 4.95LYY139 pKa = 9.16TINDD143 pKa = 3.5PTTDD147 pKa = 3.2VGGVQPTPTVVSTEE161 pKa = 3.95EE162 pKa = 3.94GAVAIIDD169 pKa = 3.8AQPVPEE175 pKa = 4.45RR176 pKa = 11.84PVQVFYY182 pKa = 11.26DD183 pKa = 4.21PNPVTDD189 pKa = 3.39SSVNIFPAYY198 pKa = 6.87PTTSTDD204 pKa = 2.67VSIFVDD210 pKa = 3.74SFSSDD215 pKa = 3.5VIGGFDD221 pKa = 4.42EE222 pKa = 5.7IPLQRR227 pKa = 11.84LDD229 pKa = 4.02FSEE232 pKa = 5.67LDD234 pKa = 3.58IEE236 pKa = 5.38DD237 pKa = 3.82IPAASTPVQKK247 pKa = 10.92VEE249 pKa = 4.04AALSRR254 pKa = 11.84AKK256 pKa = 10.29SLYY259 pKa = 9.9SRR261 pKa = 11.84YY262 pKa = 9.34FKK264 pKa = 10.36QVPVRR269 pKa = 11.84SSRR272 pKa = 11.84FLTQPSSLVQFEE284 pKa = 4.64FEE286 pKa = 4.14NPAFEE291 pKa = 4.52SEE293 pKa = 3.71ISIQVEE299 pKa = 3.48RR300 pKa = 11.84DD301 pKa = 3.18LAQVTAAPDD310 pKa = 3.23AEE312 pKa = 4.26FADD315 pKa = 4.9VITLHH320 pKa = 6.76RR321 pKa = 11.84PQLSSVEE328 pKa = 3.98GTVRR332 pKa = 11.84VSRR335 pKa = 11.84LGEE338 pKa = 3.99TGTMSTRR345 pKa = 11.84SGLIIGQRR353 pKa = 11.84VHH355 pKa = 7.27FYY357 pKa = 10.82HH358 pKa = 7.41DD359 pKa = 3.55LSEE362 pKa = 4.25IQPAEE367 pKa = 4.27NIEE370 pKa = 3.95MAILGEE376 pKa = 4.03HH377 pKa = 6.37SGLNTVVDD385 pKa = 5.58DD386 pKa = 4.17ILSSTVVDD394 pKa = 3.75PTNATNIQYY403 pKa = 10.4PEE405 pKa = 4.06DD406 pKa = 5.35AILDD410 pKa = 4.13DD411 pKa = 5.13LDD413 pKa = 4.28EE414 pKa = 5.68DD415 pKa = 4.08FTNTHH420 pKa = 6.26LVVTNTAEE428 pKa = 4.1NEE430 pKa = 4.11EE431 pKa = 4.37SDD433 pKa = 4.18LVPTIVLRR441 pKa = 11.84NSSTTVTADD450 pKa = 2.9INNGVYY456 pKa = 10.22VSYY459 pKa = 10.93PNTLNNVFTPQIISPASILPQYY481 pKa = 10.47VIEE484 pKa = 4.27TFHH487 pKa = 7.75DD488 pKa = 4.76FFLTPDD494 pKa = 4.04LYY496 pKa = 9.76PRR498 pKa = 11.84KK499 pKa = 9.51RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84IDD504 pKa = 3.29FFF506 pKa = 5.79
MM1 pKa = 7.07EE2 pKa = 5.43AKK4 pKa = 9.35TRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.5RR9 pKa = 11.84AAPDD13 pKa = 2.91QLYY16 pKa = 9.56RR17 pKa = 11.84HH18 pKa = 6.65CLQGGDD24 pKa = 5.15CIPDD28 pKa = 3.33VQNKK32 pKa = 9.5YY33 pKa = 7.08EE34 pKa = 4.1QKK36 pKa = 8.24TWADD40 pKa = 3.39VLLKK44 pKa = 10.51IFGSLLYY51 pKa = 10.37FGNLGIGTGRR61 pKa = 11.84GSGGSLGYY69 pKa = 10.22RR70 pKa = 11.84PLGTTRR76 pKa = 11.84VGEE79 pKa = 4.19VAPITPARR87 pKa = 11.84PSILIDD93 pKa = 3.34AVGPTDD99 pKa = 4.03VVPIDD104 pKa = 3.76ASTPAIVPLAEE115 pKa = 4.29GTLDD119 pKa = 3.39TGFIAPDD126 pKa = 3.64AGPGVGVEE134 pKa = 4.06EE135 pKa = 4.93LEE137 pKa = 4.95LYY139 pKa = 9.16TINDD143 pKa = 3.5PTTDD147 pKa = 3.2VGGVQPTPTVVSTEE161 pKa = 3.95EE162 pKa = 3.94GAVAIIDD169 pKa = 3.8AQPVPEE175 pKa = 4.45RR176 pKa = 11.84PVQVFYY182 pKa = 11.26DD183 pKa = 4.21PNPVTDD189 pKa = 3.39SSVNIFPAYY198 pKa = 6.87PTTSTDD204 pKa = 2.67VSIFVDD210 pKa = 3.74SFSSDD215 pKa = 3.5VIGGFDD221 pKa = 4.42EE222 pKa = 5.7IPLQRR227 pKa = 11.84LDD229 pKa = 4.02FSEE232 pKa = 5.67LDD234 pKa = 3.58IEE236 pKa = 5.38DD237 pKa = 3.82IPAASTPVQKK247 pKa = 10.92VEE249 pKa = 4.04AALSRR254 pKa = 11.84AKK256 pKa = 10.29SLYY259 pKa = 9.9SRR261 pKa = 11.84YY262 pKa = 9.34FKK264 pKa = 10.36QVPVRR269 pKa = 11.84SSRR272 pKa = 11.84FLTQPSSLVQFEE284 pKa = 4.64FEE286 pKa = 4.14NPAFEE291 pKa = 4.52SEE293 pKa = 3.71ISIQVEE299 pKa = 3.48RR300 pKa = 11.84DD301 pKa = 3.18LAQVTAAPDD310 pKa = 3.23AEE312 pKa = 4.26FADD315 pKa = 4.9VITLHH320 pKa = 6.76RR321 pKa = 11.84PQLSSVEE328 pKa = 3.98GTVRR332 pKa = 11.84VSRR335 pKa = 11.84LGEE338 pKa = 3.99TGTMSTRR345 pKa = 11.84SGLIIGQRR353 pKa = 11.84VHH355 pKa = 7.27FYY357 pKa = 10.82HH358 pKa = 7.41DD359 pKa = 3.55LSEE362 pKa = 4.25IQPAEE367 pKa = 4.27NIEE370 pKa = 3.95MAILGEE376 pKa = 4.03HH377 pKa = 6.37SGLNTVVDD385 pKa = 5.58DD386 pKa = 4.17ILSSTVVDD394 pKa = 3.75PTNATNIQYY403 pKa = 10.4PEE405 pKa = 4.06DD406 pKa = 5.35AILDD410 pKa = 4.13DD411 pKa = 5.13LDD413 pKa = 4.28EE414 pKa = 5.68DD415 pKa = 4.08FTNTHH420 pKa = 6.26LVVTNTAEE428 pKa = 4.1NEE430 pKa = 4.11EE431 pKa = 4.37SDD433 pKa = 4.18LVPTIVLRR441 pKa = 11.84NSSTTVTADD450 pKa = 2.9INNGVYY456 pKa = 10.22VSYY459 pKa = 10.93PNTLNNVFTPQIISPASILPQYY481 pKa = 10.47VIEE484 pKa = 4.27TFHH487 pKa = 7.75DD488 pKa = 4.76FFLTPDD494 pKa = 4.04LYY496 pKa = 9.76PRR498 pKa = 11.84KK499 pKa = 9.51RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84IDD504 pKa = 3.29FFF506 pKa = 5.79
Molecular weight: 55.11 kDa
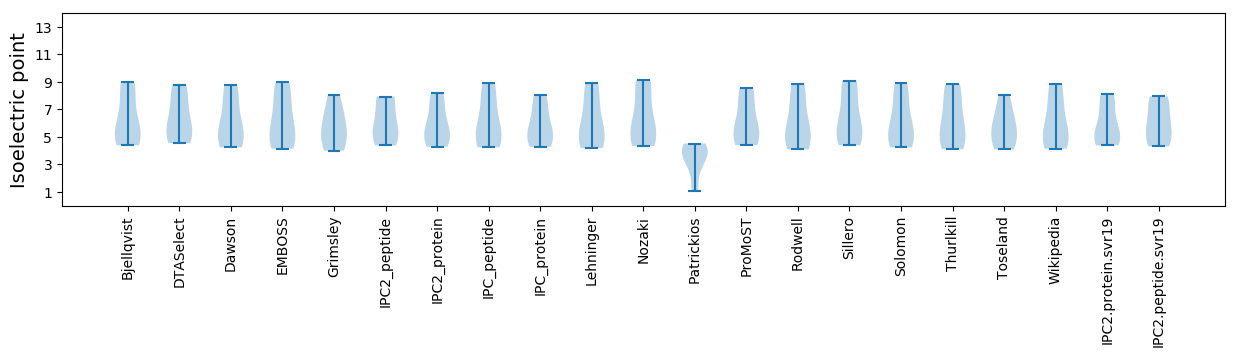
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166FK53|A0A166FK53_9PAPI Minor capsid protein L2 OS=Human papillomavirus 157 OX=2259331 GN=L2 PE=3 SV=1
MM1 pKa = 7.32EE2 pKa = 4.51TQQTLAEE9 pKa = 4.16RR10 pKa = 11.84FAAQQEE16 pKa = 4.55TQMTLIEE23 pKa = 4.15QEE25 pKa = 4.28STNLRR30 pKa = 11.84DD31 pKa = 4.91HH32 pKa = 5.8ITYY35 pKa = 9.13WDD37 pKa = 3.62SVRR40 pKa = 11.84QEE42 pKa = 3.96NLLAFYY48 pKa = 10.33AKK50 pKa = 10.41KK51 pKa = 10.28EE52 pKa = 4.4GYY54 pKa = 7.23THH56 pKa = 7.86LGLQPLAALAVSEE69 pKa = 4.36YY70 pKa = 10.15RR71 pKa = 11.84CKK73 pKa = 10.71EE74 pKa = 3.72AMKK77 pKa = 10.15MKK79 pKa = 9.73MLLISLSKK87 pKa = 10.08SAYY90 pKa = 9.94GNEE93 pKa = 3.88PWTLSDD99 pKa = 4.6VSAEE103 pKa = 4.84LINTSPKK110 pKa = 10.65NCFKK114 pKa = 11.08KK115 pKa = 10.49NAFTVTVYY123 pKa = 10.67FDD125 pKa = 3.46NDD127 pKa = 3.3DD128 pKa = 4.11RR129 pKa = 11.84NTFPYY134 pKa = 10.05VCWEE138 pKa = 3.98SIYY141 pKa = 10.87YY142 pKa = 9.67QDD144 pKa = 5.72EE145 pKa = 3.79NSEE148 pKa = 3.91WHH150 pKa = 6.04KK151 pKa = 11.02VAGEE155 pKa = 3.89VDD157 pKa = 3.77VNGLYY162 pKa = 10.49FKK164 pKa = 10.57EE165 pKa = 3.91ITGDD169 pKa = 2.81ISYY172 pKa = 9.87FQLFQPDD179 pKa = 3.5AEE181 pKa = 4.77KK182 pKa = 10.95YY183 pKa = 8.49GHH185 pKa = 6.14SGHH188 pKa = 6.59WSVRR192 pKa = 11.84YY193 pKa = 9.74KK194 pKa = 10.93NQILSTSVTSSSRR207 pKa = 11.84AAAEE211 pKa = 4.0PEE213 pKa = 4.15SRR215 pKa = 11.84TEE217 pKa = 3.98RR218 pKa = 11.84PPTHH222 pKa = 7.22PISVPKK228 pKa = 9.87TPRR231 pKa = 11.84KK232 pKa = 9.55RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84QTDD238 pKa = 2.87EE239 pKa = 3.69DD240 pKa = 4.38TNRR243 pKa = 11.84EE244 pKa = 4.36SPTSTSSGLRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84GSEE261 pKa = 3.75QGEE264 pKa = 4.55SSTDD268 pKa = 3.09GATTRR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84SRR278 pKa = 11.84TRR280 pKa = 11.84RR281 pKa = 11.84ISDD284 pKa = 3.76GSAVSPEE291 pKa = 4.07QVGSRR296 pKa = 11.84TEE298 pKa = 3.93SVPRR302 pKa = 11.84EE303 pKa = 3.62GLGRR307 pKa = 11.84IEE309 pKa = 4.25RR310 pKa = 11.84LQEE313 pKa = 3.72EE314 pKa = 4.49ARR316 pKa = 11.84DD317 pKa = 3.77PPVILIQGCANNLKK331 pKa = 10.35CFRR334 pKa = 11.84NRR336 pKa = 11.84NHH338 pKa = 6.61HH339 pKa = 5.06RR340 pKa = 11.84HH341 pKa = 4.62KK342 pKa = 10.32HH343 pKa = 5.08LYY345 pKa = 9.19LAASTVFTWVYY356 pKa = 10.65SNNDD360 pKa = 3.17RR361 pKa = 11.84TPQGRR366 pKa = 11.84MLVSFTSNSQRR377 pKa = 11.84DD378 pKa = 3.66LFLRR382 pKa = 11.84TVTIPKK388 pKa = 8.92GCRR391 pKa = 11.84YY392 pKa = 10.29CFGTLDD398 pKa = 3.85CLL400 pKa = 4.18
MM1 pKa = 7.32EE2 pKa = 4.51TQQTLAEE9 pKa = 4.16RR10 pKa = 11.84FAAQQEE16 pKa = 4.55TQMTLIEE23 pKa = 4.15QEE25 pKa = 4.28STNLRR30 pKa = 11.84DD31 pKa = 4.91HH32 pKa = 5.8ITYY35 pKa = 9.13WDD37 pKa = 3.62SVRR40 pKa = 11.84QEE42 pKa = 3.96NLLAFYY48 pKa = 10.33AKK50 pKa = 10.41KK51 pKa = 10.28EE52 pKa = 4.4GYY54 pKa = 7.23THH56 pKa = 7.86LGLQPLAALAVSEE69 pKa = 4.36YY70 pKa = 10.15RR71 pKa = 11.84CKK73 pKa = 10.71EE74 pKa = 3.72AMKK77 pKa = 10.15MKK79 pKa = 9.73MLLISLSKK87 pKa = 10.08SAYY90 pKa = 9.94GNEE93 pKa = 3.88PWTLSDD99 pKa = 4.6VSAEE103 pKa = 4.84LINTSPKK110 pKa = 10.65NCFKK114 pKa = 11.08KK115 pKa = 10.49NAFTVTVYY123 pKa = 10.67FDD125 pKa = 3.46NDD127 pKa = 3.3DD128 pKa = 4.11RR129 pKa = 11.84NTFPYY134 pKa = 10.05VCWEE138 pKa = 3.98SIYY141 pKa = 10.87YY142 pKa = 9.67QDD144 pKa = 5.72EE145 pKa = 3.79NSEE148 pKa = 3.91WHH150 pKa = 6.04KK151 pKa = 11.02VAGEE155 pKa = 3.89VDD157 pKa = 3.77VNGLYY162 pKa = 10.49FKK164 pKa = 10.57EE165 pKa = 3.91ITGDD169 pKa = 2.81ISYY172 pKa = 9.87FQLFQPDD179 pKa = 3.5AEE181 pKa = 4.77KK182 pKa = 10.95YY183 pKa = 8.49GHH185 pKa = 6.14SGHH188 pKa = 6.59WSVRR192 pKa = 11.84YY193 pKa = 9.74KK194 pKa = 10.93NQILSTSVTSSSRR207 pKa = 11.84AAAEE211 pKa = 4.0PEE213 pKa = 4.15SRR215 pKa = 11.84TEE217 pKa = 3.98RR218 pKa = 11.84PPTHH222 pKa = 7.22PISVPKK228 pKa = 9.87TPRR231 pKa = 11.84KK232 pKa = 9.55RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84QTDD238 pKa = 2.87EE239 pKa = 3.69DD240 pKa = 4.38TNRR243 pKa = 11.84EE244 pKa = 4.36SPTSTSSGLRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84GSEE261 pKa = 3.75QGEE264 pKa = 4.55SSTDD268 pKa = 3.09GATTRR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84SRR278 pKa = 11.84TRR280 pKa = 11.84RR281 pKa = 11.84ISDD284 pKa = 3.76GSAVSPEE291 pKa = 4.07QVGSRR296 pKa = 11.84TEE298 pKa = 3.93SVPRR302 pKa = 11.84EE303 pKa = 3.62GLGRR307 pKa = 11.84IEE309 pKa = 4.25RR310 pKa = 11.84LQEE313 pKa = 3.72EE314 pKa = 4.49ARR316 pKa = 11.84DD317 pKa = 3.77PPVILIQGCANNLKK331 pKa = 10.35CFRR334 pKa = 11.84NRR336 pKa = 11.84NHH338 pKa = 6.61HH339 pKa = 5.06RR340 pKa = 11.84HH341 pKa = 4.62KK342 pKa = 10.32HH343 pKa = 5.08LYY345 pKa = 9.19LAASTVFTWVYY356 pKa = 10.65SNNDD360 pKa = 3.17RR361 pKa = 11.84TPQGRR366 pKa = 11.84MLVSFTSNSQRR377 pKa = 11.84DD378 pKa = 3.66LFLRR382 pKa = 11.84TVTIPKK388 pKa = 8.92GCRR391 pKa = 11.84YY392 pKa = 10.29CFGTLDD398 pKa = 3.85CLL400 pKa = 4.18
Molecular weight: 45.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2248 |
93 |
601 |
374.7 |
42.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.139 ± 0.317 | 2.491 ± 0.784 |
6.717 ± 0.512 | 6.228 ± 0.408 |
4.982 ± 0.467 | 5.071 ± 0.761 |
1.957 ± 0.273 | 5.205 ± 0.468 |
5.071 ± 0.868 | 8.541 ± 0.403 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.36 | 5.16 ± 0.607 |
5.427 ± 0.86 | 4.226 ± 0.132 |
5.649 ± 0.927 | 7.518 ± 0.689 |
6.895 ± 0.605 | 6.361 ± 0.648 |
1.201 ± 0.224 | 3.514 ± 0.227 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
