
Termite gut associated microvirus 1
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
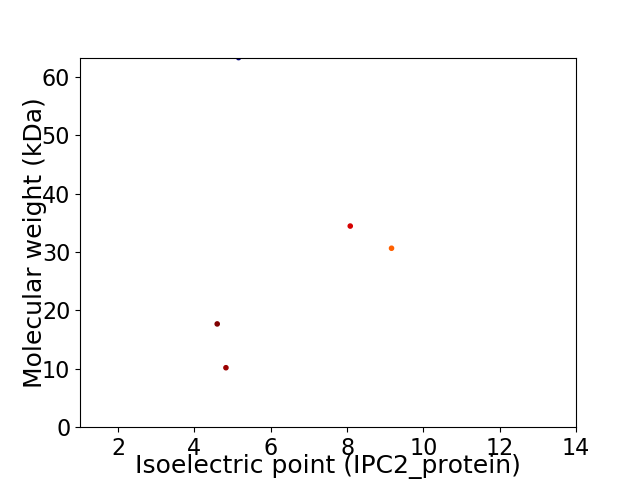
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
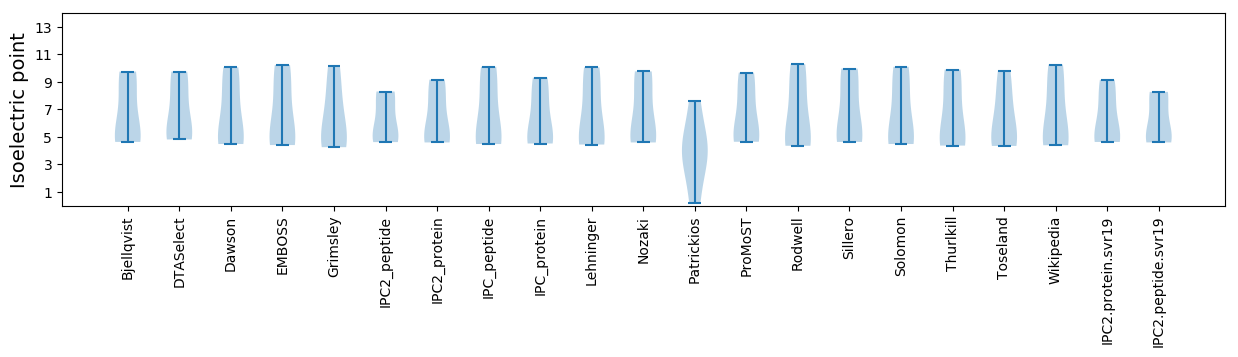
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G3IST8|A0A3G3IST8_9VIRU DNA pilot protein OS=Termite gut associated microvirus 1 OX=2486188 PE=4 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.46QSVSVDD9 pKa = 3.16DD10 pKa = 3.89VVIYY14 pKa = 9.38SHH16 pKa = 6.69YY17 pKa = 9.68YY18 pKa = 7.31PRR20 pKa = 11.84RR21 pKa = 11.84VPGITFTEE29 pKa = 4.17PTMAQQHH36 pKa = 5.53FRR38 pKa = 11.84DD39 pKa = 3.7EE40 pKa = 4.36CDD42 pKa = 2.78INTIMSRR49 pKa = 11.84YY50 pKa = 8.43EE51 pKa = 3.76RR52 pKa = 11.84TGVLVDD58 pKa = 3.95PLQQRR63 pKa = 11.84TAKK66 pKa = 10.52PMFDD70 pKa = 4.39DD71 pKa = 5.17FSVIPDD77 pKa = 3.45YY78 pKa = 11.48QEE80 pKa = 3.72AQNAIIQARR89 pKa = 11.84YY90 pKa = 9.91LFDD93 pKa = 4.27NLPASIRR100 pKa = 11.84KK101 pKa = 9.16RR102 pKa = 11.84FNNDD106 pKa = 2.89PAQLLDD112 pKa = 5.15FMADD116 pKa = 3.19DD117 pKa = 4.44SNFEE121 pKa = 3.91EE122 pKa = 4.99AVALGIVAKK131 pKa = 10.19PLEE134 pKa = 4.61TPPKK138 pKa = 10.4SEE140 pKa = 5.0DD141 pKa = 3.38KK142 pKa = 10.47PSEE145 pKa = 4.08PEE147 pKa = 4.1VVNKK151 pKa = 10.55DD152 pKa = 3.41VTPP155 pKa = 3.96
MM1 pKa = 7.53SKK3 pKa = 10.46QSVSVDD9 pKa = 3.16DD10 pKa = 3.89VVIYY14 pKa = 9.38SHH16 pKa = 6.69YY17 pKa = 9.68YY18 pKa = 7.31PRR20 pKa = 11.84RR21 pKa = 11.84VPGITFTEE29 pKa = 4.17PTMAQQHH36 pKa = 5.53FRR38 pKa = 11.84DD39 pKa = 3.7EE40 pKa = 4.36CDD42 pKa = 2.78INTIMSRR49 pKa = 11.84YY50 pKa = 8.43EE51 pKa = 3.76RR52 pKa = 11.84TGVLVDD58 pKa = 3.95PLQQRR63 pKa = 11.84TAKK66 pKa = 10.52PMFDD70 pKa = 4.39DD71 pKa = 5.17FSVIPDD77 pKa = 3.45YY78 pKa = 11.48QEE80 pKa = 3.72AQNAIIQARR89 pKa = 11.84YY90 pKa = 9.91LFDD93 pKa = 4.27NLPASIRR100 pKa = 11.84KK101 pKa = 9.16RR102 pKa = 11.84FNNDD106 pKa = 2.89PAQLLDD112 pKa = 5.15FMADD116 pKa = 3.19DD117 pKa = 4.44SNFEE121 pKa = 3.91EE122 pKa = 4.99AVALGIVAKK131 pKa = 10.19PLEE134 pKa = 4.61TPPKK138 pKa = 10.4SEE140 pKa = 5.0DD141 pKa = 3.38KK142 pKa = 10.47PSEE145 pKa = 4.08PEE147 pKa = 4.1VVNKK151 pKa = 10.55DD152 pKa = 3.41VTPP155 pKa = 3.96
Molecular weight: 17.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G3IST8|A0A3G3IST8_9VIRU DNA pilot protein OS=Termite gut associated microvirus 1 OX=2486188 PE=4 SV=1
MM1 pKa = 6.88QVASARR7 pKa = 11.84EE8 pKa = 3.61QMAFQEE14 pKa = 4.01RR15 pKa = 11.84MFRR18 pKa = 11.84NRR20 pKa = 11.84HH21 pKa = 4.18TYY23 pKa = 8.73EE24 pKa = 3.87VEE26 pKa = 3.73DD27 pKa = 4.28LKK29 pKa = 11.19RR30 pKa = 11.84AGLNPILSAHH40 pKa = 6.45SGGSTPSGAMVGGMEE55 pKa = 4.35NVGEE59 pKa = 4.2SAVRR63 pKa = 11.84GFSAAQQASIAKK75 pKa = 9.69DD76 pKa = 3.21QLALNKK82 pKa = 10.28SKK84 pKa = 11.08LEE86 pKa = 3.92SEE88 pKa = 4.25IAVNSATAQRR98 pKa = 11.84QQSEE102 pKa = 4.32SALIKK107 pKa = 10.36SQVDD111 pKa = 3.06SGYY114 pKa = 9.97YY115 pKa = 9.31PSLVGLNNASAQEE128 pKa = 3.87RR129 pKa = 11.84AAQIPLIEE137 pKa = 4.35SSVRR141 pKa = 11.84LNGQRR146 pKa = 11.84ILEE149 pKa = 4.19SDD151 pKa = 3.35ARR153 pKa = 11.84VQQLRR158 pKa = 11.84KK159 pKa = 10.02DD160 pKa = 3.08ISLIEE165 pKa = 3.81EE166 pKa = 4.76RR167 pKa = 11.84IRR169 pKa = 11.84TEE171 pKa = 3.45RR172 pKa = 11.84SLRR175 pKa = 11.84ALNAANTQLTYY186 pKa = 11.08LRR188 pKa = 11.84GALTEE193 pKa = 4.2EE194 pKa = 4.49EE195 pKa = 4.59TRR197 pKa = 11.84TQQNITSLQGYY208 pKa = 8.48LAEE211 pKa = 4.38AQRR214 pKa = 11.84LGIPEE219 pKa = 4.25KK220 pKa = 10.72LIGYY224 pKa = 9.07IKK226 pKa = 10.49YY227 pKa = 10.64SNPRR231 pKa = 11.84LQSLLSQGLTKK242 pKa = 10.47QEE244 pKa = 4.71LYY246 pKa = 10.3PFAIGTGQSLFNSAVEE262 pKa = 4.23KK263 pKa = 10.86ARR265 pKa = 11.84SNLPNRR271 pKa = 11.84VNRR274 pKa = 11.84SHH276 pKa = 7.02KK277 pKa = 10.31RR278 pKa = 3.27
MM1 pKa = 6.88QVASARR7 pKa = 11.84EE8 pKa = 3.61QMAFQEE14 pKa = 4.01RR15 pKa = 11.84MFRR18 pKa = 11.84NRR20 pKa = 11.84HH21 pKa = 4.18TYY23 pKa = 8.73EE24 pKa = 3.87VEE26 pKa = 3.73DD27 pKa = 4.28LKK29 pKa = 11.19RR30 pKa = 11.84AGLNPILSAHH40 pKa = 6.45SGGSTPSGAMVGGMEE55 pKa = 4.35NVGEE59 pKa = 4.2SAVRR63 pKa = 11.84GFSAAQQASIAKK75 pKa = 9.69DD76 pKa = 3.21QLALNKK82 pKa = 10.28SKK84 pKa = 11.08LEE86 pKa = 3.92SEE88 pKa = 4.25IAVNSATAQRR98 pKa = 11.84QQSEE102 pKa = 4.32SALIKK107 pKa = 10.36SQVDD111 pKa = 3.06SGYY114 pKa = 9.97YY115 pKa = 9.31PSLVGLNNASAQEE128 pKa = 3.87RR129 pKa = 11.84AAQIPLIEE137 pKa = 4.35SSVRR141 pKa = 11.84LNGQRR146 pKa = 11.84ILEE149 pKa = 4.19SDD151 pKa = 3.35ARR153 pKa = 11.84VQQLRR158 pKa = 11.84KK159 pKa = 10.02DD160 pKa = 3.08ISLIEE165 pKa = 3.81EE166 pKa = 4.76RR167 pKa = 11.84IRR169 pKa = 11.84TEE171 pKa = 3.45RR172 pKa = 11.84SLRR175 pKa = 11.84ALNAANTQLTYY186 pKa = 11.08LRR188 pKa = 11.84GALTEE193 pKa = 4.2EE194 pKa = 4.49EE195 pKa = 4.59TRR197 pKa = 11.84TQQNITSLQGYY208 pKa = 8.48LAEE211 pKa = 4.38AQRR214 pKa = 11.84LGIPEE219 pKa = 4.25KK220 pKa = 10.72LIGYY224 pKa = 9.07IKK226 pKa = 10.49YY227 pKa = 10.64SNPRR231 pKa = 11.84LQSLLSQGLTKK242 pKa = 10.47QEE244 pKa = 4.71LYY246 pKa = 10.3PFAIGTGQSLFNSAVEE262 pKa = 4.23KK263 pKa = 10.86ARR265 pKa = 11.84SNLPNRR271 pKa = 11.84VNRR274 pKa = 11.84SHH276 pKa = 7.02KK277 pKa = 10.31RR278 pKa = 3.27
Molecular weight: 30.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1384 |
91 |
567 |
276.8 |
31.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.587 ± 0.93 | 1.301 ± 0.612 |
6.431 ± 1.152 | 5.13 ± 1.047 |
4.408 ± 0.889 | 6.72 ± 1.017 |
2.24 ± 0.567 | 5.347 ± 0.185 |
3.251 ± 0.711 | 7.442 ± 0.825 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.806 ± 0.299 | 4.697 ± 0.451 |
5.708 ± 0.952 | 5.202 ± 0.963 |
7.514 ± 1.031 | 7.298 ± 0.897 |
5.058 ± 0.522 | 6.792 ± 0.789 |
1.156 ± 0.463 | 4.913 ± 0.636 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
