
Eubacteriaceae bacterium CHKCI005
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; unclassified Eubacteriaceae
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
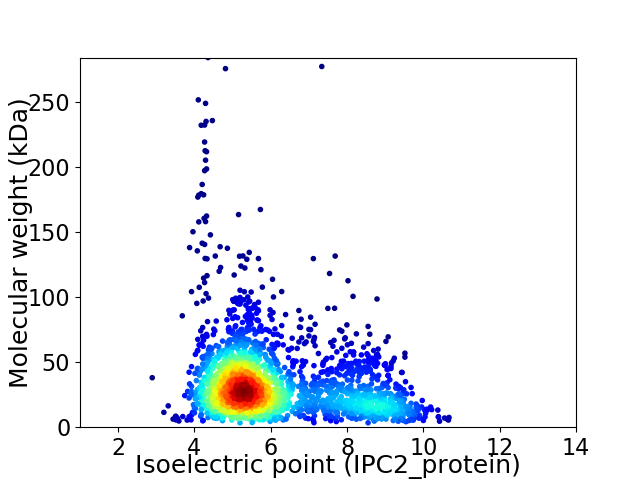
Virtual 2D-PAGE plot for 2248 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143ZXI7|A0A143ZXI7_9FIRM Uncharacterized protein OS=Eubacteriaceae bacterium CHKCI005 OX=1780381 GN=BN3661_01470 PE=4 SV=1
MM1 pKa = 7.14STHH4 pKa = 5.64KK5 pKa = 10.23HH6 pKa = 5.71INTICCISLVAVLLITILFLGFGSTGIVAVSKK38 pKa = 10.77ALGYY42 pKa = 8.58EE43 pKa = 3.71EE44 pKa = 5.74RR45 pKa = 11.84LFDD48 pKa = 3.51TSRR51 pKa = 11.84VHH53 pKa = 6.27TLDD56 pKa = 3.04IVMDD60 pKa = 3.52NWDD63 pKa = 4.13EE64 pKa = 4.96FVDD67 pKa = 3.47NCQNEE72 pKa = 4.81EE73 pKa = 4.25YY74 pKa = 10.31VSCSVVIDD82 pKa = 3.57NEE84 pKa = 4.27SFKK87 pKa = 11.29NVAIRR92 pKa = 11.84AKK94 pKa = 10.67GNTSLSSVAAYY105 pKa = 10.51GNDD108 pKa = 3.09RR109 pKa = 11.84YY110 pKa = 11.01SFKK113 pKa = 10.67IEE115 pKa = 3.6FDD117 pKa = 3.18HH118 pKa = 8.53YY119 pKa = 11.24EE120 pKa = 4.2DD121 pKa = 3.77GKK123 pKa = 11.22SYY125 pKa = 10.87HH126 pKa = 6.77GLDD129 pKa = 3.58KK130 pKa = 11.3LSLNNIIQDD139 pKa = 3.55NTYY142 pKa = 9.84MKK144 pKa = 10.54DD145 pKa = 3.59YY146 pKa = 10.88LCYY149 pKa = 10.87QMMGAFGVDD158 pKa = 3.62SPLCSYY164 pKa = 10.78IYY166 pKa = 9.4ITVNGEE172 pKa = 3.56DD173 pKa = 3.09WGLYY177 pKa = 9.44LAVEE181 pKa = 4.5GVEE184 pKa = 4.28EE185 pKa = 4.84SFLQRR190 pKa = 11.84NYY192 pKa = 10.87GSDD195 pKa = 3.4YY196 pKa = 11.49GEE198 pKa = 4.55LYY200 pKa = 10.75KK201 pKa = 10.4PDD203 pKa = 3.78STSMGGGRR211 pKa = 11.84GNGGNFDD218 pKa = 3.57FDD220 pKa = 3.74RR221 pKa = 11.84FQEE224 pKa = 4.2EE225 pKa = 4.35QEE227 pKa = 4.46GQDD230 pKa = 3.41GTTDD234 pKa = 4.18DD235 pKa = 5.59DD236 pKa = 4.12DD237 pKa = 4.31TTGSGSGDD245 pKa = 3.33NTQPTPPDD253 pKa = 4.12GNWGDD258 pKa = 3.69NFSGNMPGPGEE269 pKa = 4.11FPGGSSDD276 pKa = 5.71DD277 pKa = 3.99GMPDD281 pKa = 2.92MGDD284 pKa = 3.07FPGFSEE290 pKa = 5.0DD291 pKa = 3.85DD292 pKa = 3.69SSGEE296 pKa = 4.07TQGNIPAIKK305 pKa = 10.04DD306 pKa = 3.53FTGGSSPEE314 pKa = 4.14GDD316 pKa = 3.95SEE318 pKa = 4.73DD319 pKa = 4.28GPGNMEE325 pKa = 4.4GPGGMGDD332 pKa = 3.5SDD334 pKa = 3.83VSLIYY339 pKa = 10.59TDD341 pKa = 5.42DD342 pKa = 5.38DD343 pKa = 4.19YY344 pKa = 12.13DD345 pKa = 3.81SYY347 pKa = 12.21RR348 pKa = 11.84NIFDD352 pKa = 3.6NAKK355 pKa = 9.47TDD357 pKa = 3.1ITDD360 pKa = 3.29NDD362 pKa = 3.82KK363 pKa = 11.54DD364 pKa = 3.89RR365 pKa = 11.84LIASLKK371 pKa = 10.16QLGDD375 pKa = 3.51HH376 pKa = 6.08QNISDD381 pKa = 3.73IVDD384 pKa = 2.99IDD386 pKa = 3.42EE387 pKa = 4.01VLRR390 pKa = 11.84YY391 pKa = 8.68FVVHH395 pKa = 6.37NFVCNFDD402 pKa = 4.16SYY404 pKa = 9.87TGSMIHH410 pKa = 6.56NYY412 pKa = 9.82YY413 pKa = 10.48LYY415 pKa = 10.83EE416 pKa = 4.34EE417 pKa = 5.32DD418 pKa = 4.64GQLSMIPWDD427 pKa = 3.59YY428 pKa = 11.69NLAFGGFMSMGGATSLVNYY447 pKa = 9.6PIDD450 pKa = 3.82TPVSGGTVDD459 pKa = 4.2SRR461 pKa = 11.84PMLAWIFQNDD471 pKa = 4.36EE472 pKa = 4.05YY473 pKa = 10.95TQMYY477 pKa = 9.52HH478 pKa = 7.8DD479 pKa = 4.97YY480 pKa = 9.56FAQFIADD487 pKa = 3.98TFDD490 pKa = 3.63NGCFASMMDD499 pKa = 3.94KK500 pKa = 10.51VVEE503 pKa = 4.64LISPYY508 pKa = 8.05VQKK511 pKa = 11.01DD512 pKa = 3.23PTKK515 pKa = 8.66FCTYY519 pKa = 10.71EE520 pKa = 3.82EE521 pKa = 4.72FEE523 pKa = 4.45TGISTLEE530 pKa = 3.87QFCTLRR536 pKa = 11.84AQSISGQLDD545 pKa = 3.31GSIPSTSDD553 pKa = 3.02GQSSDD558 pKa = 3.16SSSLIDD564 pKa = 4.48ASQLSIDD571 pKa = 4.0AMGSMGIGGGSGNGHH586 pKa = 5.37GQMNHH591 pKa = 6.04SDD593 pKa = 3.74EE594 pKa = 4.56DD595 pKa = 4.46TNPSNSGDD603 pKa = 3.6IGDD606 pKa = 4.35IPQNNQGDD614 pKa = 4.97FISLLNGTPLSSSSDD629 pKa = 3.21ALFASTSSAFSVSPAAFPAAPDD651 pKa = 3.52STQGNRR657 pKa = 11.84PPSGDD662 pKa = 3.24FDD664 pKa = 4.77GGMGGMGGGPAGSPPGSTDD683 pKa = 3.13DD684 pKa = 4.05SQPSGSTSQSDD695 pKa = 3.53SSSPHH700 pKa = 5.41STNDD704 pKa = 3.3SNDD707 pKa = 3.42NGQSLPSEE715 pKa = 4.46DD716 pKa = 4.12SSPGSGSNSDD726 pKa = 3.46SDD728 pKa = 4.19STQGMQPPSDD738 pKa = 3.63NSGGGMPGQDD748 pKa = 2.99EE749 pKa = 4.31NTPNGMQNPFDD760 pKa = 3.95SQTASSQWGGMDD772 pKa = 3.31TQGWILLGISIFVLGVGLLVACLMKK797 pKa = 10.64GKK799 pKa = 9.69NWW801 pKa = 3.39
MM1 pKa = 7.14STHH4 pKa = 5.64KK5 pKa = 10.23HH6 pKa = 5.71INTICCISLVAVLLITILFLGFGSTGIVAVSKK38 pKa = 10.77ALGYY42 pKa = 8.58EE43 pKa = 3.71EE44 pKa = 5.74RR45 pKa = 11.84LFDD48 pKa = 3.51TSRR51 pKa = 11.84VHH53 pKa = 6.27TLDD56 pKa = 3.04IVMDD60 pKa = 3.52NWDD63 pKa = 4.13EE64 pKa = 4.96FVDD67 pKa = 3.47NCQNEE72 pKa = 4.81EE73 pKa = 4.25YY74 pKa = 10.31VSCSVVIDD82 pKa = 3.57NEE84 pKa = 4.27SFKK87 pKa = 11.29NVAIRR92 pKa = 11.84AKK94 pKa = 10.67GNTSLSSVAAYY105 pKa = 10.51GNDD108 pKa = 3.09RR109 pKa = 11.84YY110 pKa = 11.01SFKK113 pKa = 10.67IEE115 pKa = 3.6FDD117 pKa = 3.18HH118 pKa = 8.53YY119 pKa = 11.24EE120 pKa = 4.2DD121 pKa = 3.77GKK123 pKa = 11.22SYY125 pKa = 10.87HH126 pKa = 6.77GLDD129 pKa = 3.58KK130 pKa = 11.3LSLNNIIQDD139 pKa = 3.55NTYY142 pKa = 9.84MKK144 pKa = 10.54DD145 pKa = 3.59YY146 pKa = 10.88LCYY149 pKa = 10.87QMMGAFGVDD158 pKa = 3.62SPLCSYY164 pKa = 10.78IYY166 pKa = 9.4ITVNGEE172 pKa = 3.56DD173 pKa = 3.09WGLYY177 pKa = 9.44LAVEE181 pKa = 4.5GVEE184 pKa = 4.28EE185 pKa = 4.84SFLQRR190 pKa = 11.84NYY192 pKa = 10.87GSDD195 pKa = 3.4YY196 pKa = 11.49GEE198 pKa = 4.55LYY200 pKa = 10.75KK201 pKa = 10.4PDD203 pKa = 3.78STSMGGGRR211 pKa = 11.84GNGGNFDD218 pKa = 3.57FDD220 pKa = 3.74RR221 pKa = 11.84FQEE224 pKa = 4.2EE225 pKa = 4.35QEE227 pKa = 4.46GQDD230 pKa = 3.41GTTDD234 pKa = 4.18DD235 pKa = 5.59DD236 pKa = 4.12DD237 pKa = 4.31TTGSGSGDD245 pKa = 3.33NTQPTPPDD253 pKa = 4.12GNWGDD258 pKa = 3.69NFSGNMPGPGEE269 pKa = 4.11FPGGSSDD276 pKa = 5.71DD277 pKa = 3.99GMPDD281 pKa = 2.92MGDD284 pKa = 3.07FPGFSEE290 pKa = 5.0DD291 pKa = 3.85DD292 pKa = 3.69SSGEE296 pKa = 4.07TQGNIPAIKK305 pKa = 10.04DD306 pKa = 3.53FTGGSSPEE314 pKa = 4.14GDD316 pKa = 3.95SEE318 pKa = 4.73DD319 pKa = 4.28GPGNMEE325 pKa = 4.4GPGGMGDD332 pKa = 3.5SDD334 pKa = 3.83VSLIYY339 pKa = 10.59TDD341 pKa = 5.42DD342 pKa = 5.38DD343 pKa = 4.19YY344 pKa = 12.13DD345 pKa = 3.81SYY347 pKa = 12.21RR348 pKa = 11.84NIFDD352 pKa = 3.6NAKK355 pKa = 9.47TDD357 pKa = 3.1ITDD360 pKa = 3.29NDD362 pKa = 3.82KK363 pKa = 11.54DD364 pKa = 3.89RR365 pKa = 11.84LIASLKK371 pKa = 10.16QLGDD375 pKa = 3.51HH376 pKa = 6.08QNISDD381 pKa = 3.73IVDD384 pKa = 2.99IDD386 pKa = 3.42EE387 pKa = 4.01VLRR390 pKa = 11.84YY391 pKa = 8.68FVVHH395 pKa = 6.37NFVCNFDD402 pKa = 4.16SYY404 pKa = 9.87TGSMIHH410 pKa = 6.56NYY412 pKa = 9.82YY413 pKa = 10.48LYY415 pKa = 10.83EE416 pKa = 4.34EE417 pKa = 5.32DD418 pKa = 4.64GQLSMIPWDD427 pKa = 3.59YY428 pKa = 11.69NLAFGGFMSMGGATSLVNYY447 pKa = 9.6PIDD450 pKa = 3.82TPVSGGTVDD459 pKa = 4.2SRR461 pKa = 11.84PMLAWIFQNDD471 pKa = 4.36EE472 pKa = 4.05YY473 pKa = 10.95TQMYY477 pKa = 9.52HH478 pKa = 7.8DD479 pKa = 4.97YY480 pKa = 9.56FAQFIADD487 pKa = 3.98TFDD490 pKa = 3.63NGCFASMMDD499 pKa = 3.94KK500 pKa = 10.51VVEE503 pKa = 4.64LISPYY508 pKa = 8.05VQKK511 pKa = 11.01DD512 pKa = 3.23PTKK515 pKa = 8.66FCTYY519 pKa = 10.71EE520 pKa = 3.82EE521 pKa = 4.72FEE523 pKa = 4.45TGISTLEE530 pKa = 3.87QFCTLRR536 pKa = 11.84AQSISGQLDD545 pKa = 3.31GSIPSTSDD553 pKa = 3.02GQSSDD558 pKa = 3.16SSSLIDD564 pKa = 4.48ASQLSIDD571 pKa = 4.0AMGSMGIGGGSGNGHH586 pKa = 5.37GQMNHH591 pKa = 6.04SDD593 pKa = 3.74EE594 pKa = 4.56DD595 pKa = 4.46TNPSNSGDD603 pKa = 3.6IGDD606 pKa = 4.35IPQNNQGDD614 pKa = 4.97FISLLNGTPLSSSSDD629 pKa = 3.21ALFASTSSAFSVSPAAFPAAPDD651 pKa = 3.52STQGNRR657 pKa = 11.84PPSGDD662 pKa = 3.24FDD664 pKa = 4.77GGMGGMGGGPAGSPPGSTDD683 pKa = 3.13DD684 pKa = 4.05SQPSGSTSQSDD695 pKa = 3.53SSSPHH700 pKa = 5.41STNDD704 pKa = 3.3SNDD707 pKa = 3.42NGQSLPSEE715 pKa = 4.46DD716 pKa = 4.12SSPGSGSNSDD726 pKa = 3.46SDD728 pKa = 4.19STQGMQPPSDD738 pKa = 3.63NSGGGMPGQDD748 pKa = 2.99EE749 pKa = 4.31NTPNGMQNPFDD760 pKa = 3.95SQTASSQWGGMDD772 pKa = 3.31TQGWILLGISIFVLGVGLLVACLMKK797 pKa = 10.64GKK799 pKa = 9.69NWW801 pKa = 3.39
Molecular weight: 85.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143ZSJ1|A0A143ZSJ1_9FIRM DNA topoisomerase (ATP-hydrolyzing) OS=Eubacteriaceae bacterium CHKCI005 OX=1780381 GN=gyrB_2 PE=4 SV=1
MM1 pKa = 7.91IYY3 pKa = 9.68TPKK6 pKa = 10.18QRR8 pKa = 11.84GLMMRR13 pKa = 11.84WKK15 pKa = 10.24QNRR18 pKa = 11.84LHH20 pKa = 6.94RR21 pKa = 11.84LNLLEE26 pKa = 4.67GSVSSGKK33 pKa = 7.53TWISLVLWAFWVATMPMDD51 pKa = 3.43KK52 pKa = 10.48LYY54 pKa = 10.71DD55 pKa = 3.51VRR57 pKa = 11.84EE58 pKa = 4.42IPHH61 pKa = 6.67HH62 pKa = 6.32AQAQLPCTVGRR73 pKa = 11.84VSWVIQFCIFSCVQRR88 pKa = 11.84RR89 pKa = 11.84IFVWQEE95 pKa = 3.33SSFGRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 10.04
MM1 pKa = 7.91IYY3 pKa = 9.68TPKK6 pKa = 10.18QRR8 pKa = 11.84GLMMRR13 pKa = 11.84WKK15 pKa = 10.24QNRR18 pKa = 11.84LHH20 pKa = 6.94RR21 pKa = 11.84LNLLEE26 pKa = 4.67GSVSSGKK33 pKa = 7.53TWISLVLWAFWVATMPMDD51 pKa = 3.43KK52 pKa = 10.48LYY54 pKa = 10.71DD55 pKa = 3.51VRR57 pKa = 11.84EE58 pKa = 4.42IPHH61 pKa = 6.67HH62 pKa = 6.32AQAQLPCTVGRR73 pKa = 11.84VSWVIQFCIFSCVQRR88 pKa = 11.84RR89 pKa = 11.84IFVWQEE95 pKa = 3.33SSFGRR100 pKa = 11.84RR101 pKa = 11.84KK102 pKa = 10.04
Molecular weight: 12.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
719107 |
30 |
2541 |
319.9 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.006 ± 0.06 | 1.536 ± 0.024 |
5.99 ± 0.047 | 6.714 ± 0.047 |
3.82 ± 0.035 | 7.517 ± 0.048 |
1.882 ± 0.024 | 6.388 ± 0.051 |
5.42 ± 0.048 | 9.342 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.868 ± 0.025 | 3.718 ± 0.041 |
4.041 ± 0.039 | 4.102 ± 0.035 |
4.965 ± 0.052 | 6.431 ± 0.049 |
5.488 ± 0.051 | 7.249 ± 0.044 |
0.998 ± 0.023 | 3.525 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
