
Wenzhou narna-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.86
Get precalculated fractions of proteins
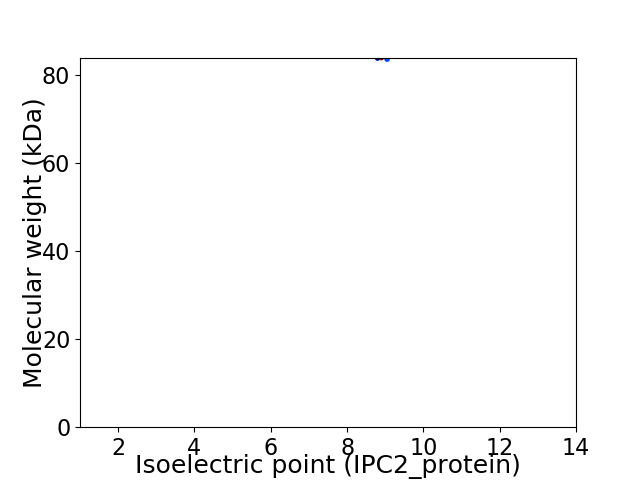
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIT8|A0A1L3KIT8_9VIRU RNA-dependent RNA polymerase OS=Wenzhou narna-like virus 7 OX=1923582 PE=4 SV=1
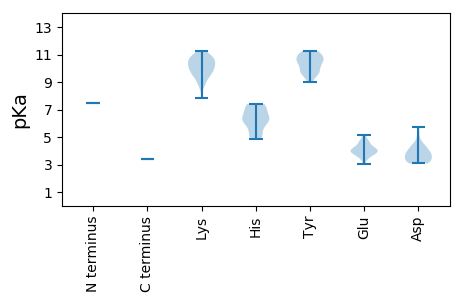
MM1 pKa = 7.44DD2 pKa = 4.22QEE4 pKa = 4.9FPCEE8 pKa = 3.84RR9 pKa = 11.84KK10 pKa = 9.17EE11 pKa = 4.64ALLDD15 pKa = 4.34HH16 pKa = 7.06INMKK20 pKa = 10.21IMSKK24 pKa = 10.14LRR26 pKa = 11.84NEE28 pKa = 4.04ARR30 pKa = 11.84HH31 pKa = 5.0KK32 pKa = 10.05RR33 pKa = 11.84KK34 pKa = 9.56RR35 pKa = 11.84AIGFAYY41 pKa = 10.38SIQNLKK47 pKa = 10.55YY48 pKa = 10.73LITFEE53 pKa = 4.57PKK55 pKa = 10.52DD56 pKa = 4.07LIKK59 pKa = 10.4STLEE63 pKa = 3.44KK64 pKa = 10.66HH65 pKa = 6.19RR66 pKa = 11.84ADD68 pKa = 3.24LSKK71 pKa = 10.63PIGGLDD77 pKa = 3.61PQIKK81 pKa = 9.25EE82 pKa = 4.11AYY84 pKa = 10.13DD85 pKa = 4.14FILQKK90 pKa = 10.93FMDD93 pKa = 5.75FIASNASEE101 pKa = 3.69IDD103 pKa = 3.73VFPEE107 pKa = 3.82PTEE110 pKa = 4.22SRR112 pKa = 11.84CHH114 pKa = 6.0SKK116 pKa = 9.07TGKK119 pKa = 9.6SFKK122 pKa = 9.87EE123 pKa = 3.99AFMKK127 pKa = 9.69ATSEE131 pKa = 4.09HH132 pKa = 6.39TSLCSNYY139 pKa = 9.63HH140 pKa = 5.38IRR142 pKa = 11.84VGDD145 pKa = 3.63RR146 pKa = 11.84TLCVPDD152 pKa = 3.39TNYY155 pKa = 11.08NKK157 pKa = 10.48DD158 pKa = 3.12FNFAHH163 pKa = 7.25GDD165 pKa = 3.61CLLNNTVKK173 pKa = 10.6AVALVEE179 pKa = 4.01PLKK182 pKa = 11.06VRR184 pKa = 11.84VITKK188 pKa = 8.8EE189 pKa = 3.81AAWTHH194 pKa = 4.87MLKK197 pKa = 10.47PIQKK201 pKa = 9.39LMHH204 pKa = 6.45SALTHH209 pKa = 6.39HH210 pKa = 7.36DD211 pKa = 3.52FGEE214 pKa = 4.47IFCLTRR220 pKa = 11.84GKK222 pKa = 9.83PYY224 pKa = 10.83EE225 pKa = 4.15NVSLGTLSDD234 pKa = 3.84SEE236 pKa = 5.14FYY238 pKa = 11.22LSGDD242 pKa = 3.89FSAATDD248 pKa = 3.89NIHH251 pKa = 6.53PHH253 pKa = 6.15FLNEE257 pKa = 3.43FRR259 pKa = 11.84DD260 pKa = 3.68RR261 pKa = 11.84MRR263 pKa = 11.84VILKK267 pKa = 7.87DD268 pKa = 3.53HH269 pKa = 6.22PMIWHH274 pKa = 7.33ALNSEE279 pKa = 4.02ARR281 pKa = 11.84NHH283 pKa = 6.25IIEE286 pKa = 4.55YY287 pKa = 9.52PLGIPPVEE295 pKa = 4.13QMRR298 pKa = 11.84GQLMGSLLSFPILCLSNLAIVLASCRR324 pKa = 11.84EE325 pKa = 4.22HH326 pKa = 7.39RR327 pKa = 11.84DD328 pKa = 3.16HH329 pKa = 7.22TIDD332 pKa = 4.83RR333 pKa = 11.84VIQSASSRR341 pKa = 11.84TGYY344 pKa = 10.55KK345 pKa = 10.2PIIEE349 pKa = 4.26KK350 pKa = 10.8VKK352 pKa = 9.5ITISEE357 pKa = 4.12NWSSTLEE364 pKa = 3.98RR365 pKa = 11.84LKK367 pKa = 10.91SSCLINGDD375 pKa = 3.89DD376 pKa = 4.51LAIRR380 pKa = 11.84TNDD383 pKa = 3.16QFLSKK388 pKa = 10.14WKK390 pKa = 10.31VLSPNAGFTLSIGKK404 pKa = 8.82NFIDD408 pKa = 4.24KK409 pKa = 10.46RR410 pKa = 11.84FYY412 pKa = 10.55TINSKK417 pKa = 10.77LFIDD421 pKa = 4.46DD422 pKa = 5.24KK423 pKa = 11.21IFSTHH428 pKa = 6.0KK429 pKa = 10.06FKK431 pKa = 11.23CSMSNKK437 pKa = 9.61GDD439 pKa = 3.8LASLGAEE446 pKa = 3.79YY447 pKa = 9.7PRR449 pKa = 11.84FFKK452 pKa = 10.85TEE454 pKa = 3.77GVQQEE459 pKa = 4.66VVDD462 pKa = 3.43KK463 pKa = 10.76HH464 pKa = 5.65NRR466 pKa = 11.84TIRR469 pKa = 11.84SMFIRR474 pKa = 11.84DD475 pKa = 3.17HH476 pKa = 6.76HH477 pKa = 6.57KK478 pKa = 10.86RR479 pKa = 11.84LMKK482 pKa = 10.39DD483 pKa = 3.19PRR485 pKa = 11.84SLEE488 pKa = 4.19VPRR491 pKa = 11.84HH492 pKa = 4.96LGGLGFGNLIPCNIMQKK509 pKa = 8.64LTHH512 pKa = 7.14LIYY515 pKa = 10.37SYY517 pKa = 11.07KK518 pKa = 8.81ITKK521 pKa = 9.52SPKK524 pKa = 9.69NFTNLLSRR532 pKa = 11.84FGLEE536 pKa = 3.07RR537 pKa = 11.84VYY539 pKa = 10.98VPRR542 pKa = 11.84GTCHH546 pKa = 7.15RR547 pKa = 11.84LKK549 pKa = 10.9LCDD552 pKa = 3.4YY553 pKa = 8.98TSYY556 pKa = 10.78IGLQFDD562 pKa = 3.85EE563 pKa = 4.71PTRR566 pKa = 11.84DD567 pKa = 3.17ISPSEE572 pKa = 3.79LPVRR576 pKa = 11.84IRR578 pKa = 11.84QYY580 pKa = 10.91IKK582 pKa = 10.63LVKK585 pKa = 9.23TKK587 pKa = 10.53QLALTDD593 pKa = 4.07RR594 pKa = 11.84WFHH597 pKa = 5.37WEE599 pKa = 3.52WVIVRR604 pKa = 11.84KK605 pKa = 9.66FDD607 pKa = 4.3LPIIRR612 pKa = 11.84QFLVDD617 pKa = 4.46LRR619 pKa = 11.84KK620 pKa = 10.07LGYY623 pKa = 10.6DD624 pKa = 3.25KK625 pKa = 11.07AISKK629 pKa = 9.99NMSYY633 pKa = 10.02IGQVIQRR640 pKa = 11.84LPTLANDD647 pKa = 3.29YY648 pKa = 11.25AKK650 pKa = 10.11FVKK653 pKa = 10.06RR654 pKa = 11.84IYY656 pKa = 9.85MKK658 pKa = 10.77RR659 pKa = 11.84LPKK662 pKa = 10.02KK663 pKa = 9.99VNGAYY668 pKa = 9.6KK669 pKa = 10.16VFRR672 pKa = 11.84DD673 pKa = 3.62SLHH676 pKa = 4.95YY677 pKa = 9.55TINTKK682 pKa = 9.53VEE684 pKa = 3.82PHH686 pKa = 6.2TEE688 pKa = 3.77LGKK691 pKa = 9.67ATVVALNYY699 pKa = 10.05AVRR702 pKa = 11.84PTLPGNTCPYY712 pKa = 10.45DD713 pKa = 3.08SWSPFDD719 pKa = 3.85GDD721 pKa = 3.1VRR723 pKa = 11.84LIVV726 pKa = 3.42
MM1 pKa = 7.44DD2 pKa = 4.22QEE4 pKa = 4.9FPCEE8 pKa = 3.84RR9 pKa = 11.84KK10 pKa = 9.17EE11 pKa = 4.64ALLDD15 pKa = 4.34HH16 pKa = 7.06INMKK20 pKa = 10.21IMSKK24 pKa = 10.14LRR26 pKa = 11.84NEE28 pKa = 4.04ARR30 pKa = 11.84HH31 pKa = 5.0KK32 pKa = 10.05RR33 pKa = 11.84KK34 pKa = 9.56RR35 pKa = 11.84AIGFAYY41 pKa = 10.38SIQNLKK47 pKa = 10.55YY48 pKa = 10.73LITFEE53 pKa = 4.57PKK55 pKa = 10.52DD56 pKa = 4.07LIKK59 pKa = 10.4STLEE63 pKa = 3.44KK64 pKa = 10.66HH65 pKa = 6.19RR66 pKa = 11.84ADD68 pKa = 3.24LSKK71 pKa = 10.63PIGGLDD77 pKa = 3.61PQIKK81 pKa = 9.25EE82 pKa = 4.11AYY84 pKa = 10.13DD85 pKa = 4.14FILQKK90 pKa = 10.93FMDD93 pKa = 5.75FIASNASEE101 pKa = 3.69IDD103 pKa = 3.73VFPEE107 pKa = 3.82PTEE110 pKa = 4.22SRR112 pKa = 11.84CHH114 pKa = 6.0SKK116 pKa = 9.07TGKK119 pKa = 9.6SFKK122 pKa = 9.87EE123 pKa = 3.99AFMKK127 pKa = 9.69ATSEE131 pKa = 4.09HH132 pKa = 6.39TSLCSNYY139 pKa = 9.63HH140 pKa = 5.38IRR142 pKa = 11.84VGDD145 pKa = 3.63RR146 pKa = 11.84TLCVPDD152 pKa = 3.39TNYY155 pKa = 11.08NKK157 pKa = 10.48DD158 pKa = 3.12FNFAHH163 pKa = 7.25GDD165 pKa = 3.61CLLNNTVKK173 pKa = 10.6AVALVEE179 pKa = 4.01PLKK182 pKa = 11.06VRR184 pKa = 11.84VITKK188 pKa = 8.8EE189 pKa = 3.81AAWTHH194 pKa = 4.87MLKK197 pKa = 10.47PIQKK201 pKa = 9.39LMHH204 pKa = 6.45SALTHH209 pKa = 6.39HH210 pKa = 7.36DD211 pKa = 3.52FGEE214 pKa = 4.47IFCLTRR220 pKa = 11.84GKK222 pKa = 9.83PYY224 pKa = 10.83EE225 pKa = 4.15NVSLGTLSDD234 pKa = 3.84SEE236 pKa = 5.14FYY238 pKa = 11.22LSGDD242 pKa = 3.89FSAATDD248 pKa = 3.89NIHH251 pKa = 6.53PHH253 pKa = 6.15FLNEE257 pKa = 3.43FRR259 pKa = 11.84DD260 pKa = 3.68RR261 pKa = 11.84MRR263 pKa = 11.84VILKK267 pKa = 7.87DD268 pKa = 3.53HH269 pKa = 6.22PMIWHH274 pKa = 7.33ALNSEE279 pKa = 4.02ARR281 pKa = 11.84NHH283 pKa = 6.25IIEE286 pKa = 4.55YY287 pKa = 9.52PLGIPPVEE295 pKa = 4.13QMRR298 pKa = 11.84GQLMGSLLSFPILCLSNLAIVLASCRR324 pKa = 11.84EE325 pKa = 4.22HH326 pKa = 7.39RR327 pKa = 11.84DD328 pKa = 3.16HH329 pKa = 7.22TIDD332 pKa = 4.83RR333 pKa = 11.84VIQSASSRR341 pKa = 11.84TGYY344 pKa = 10.55KK345 pKa = 10.2PIIEE349 pKa = 4.26KK350 pKa = 10.8VKK352 pKa = 9.5ITISEE357 pKa = 4.12NWSSTLEE364 pKa = 3.98RR365 pKa = 11.84LKK367 pKa = 10.91SSCLINGDD375 pKa = 3.89DD376 pKa = 4.51LAIRR380 pKa = 11.84TNDD383 pKa = 3.16QFLSKK388 pKa = 10.14WKK390 pKa = 10.31VLSPNAGFTLSIGKK404 pKa = 8.82NFIDD408 pKa = 4.24KK409 pKa = 10.46RR410 pKa = 11.84FYY412 pKa = 10.55TINSKK417 pKa = 10.77LFIDD421 pKa = 4.46DD422 pKa = 5.24KK423 pKa = 11.21IFSTHH428 pKa = 6.0KK429 pKa = 10.06FKK431 pKa = 11.23CSMSNKK437 pKa = 9.61GDD439 pKa = 3.8LASLGAEE446 pKa = 3.79YY447 pKa = 9.7PRR449 pKa = 11.84FFKK452 pKa = 10.85TEE454 pKa = 3.77GVQQEE459 pKa = 4.66VVDD462 pKa = 3.43KK463 pKa = 10.76HH464 pKa = 5.65NRR466 pKa = 11.84TIRR469 pKa = 11.84SMFIRR474 pKa = 11.84DD475 pKa = 3.17HH476 pKa = 6.76HH477 pKa = 6.57KK478 pKa = 10.86RR479 pKa = 11.84LMKK482 pKa = 10.39DD483 pKa = 3.19PRR485 pKa = 11.84SLEE488 pKa = 4.19VPRR491 pKa = 11.84HH492 pKa = 4.96LGGLGFGNLIPCNIMQKK509 pKa = 8.64LTHH512 pKa = 7.14LIYY515 pKa = 10.37SYY517 pKa = 11.07KK518 pKa = 8.81ITKK521 pKa = 9.52SPKK524 pKa = 9.69NFTNLLSRR532 pKa = 11.84FGLEE536 pKa = 3.07RR537 pKa = 11.84VYY539 pKa = 10.98VPRR542 pKa = 11.84GTCHH546 pKa = 7.15RR547 pKa = 11.84LKK549 pKa = 10.9LCDD552 pKa = 3.4YY553 pKa = 8.98TSYY556 pKa = 10.78IGLQFDD562 pKa = 3.85EE563 pKa = 4.71PTRR566 pKa = 11.84DD567 pKa = 3.17ISPSEE572 pKa = 3.79LPVRR576 pKa = 11.84IRR578 pKa = 11.84QYY580 pKa = 10.91IKK582 pKa = 10.63LVKK585 pKa = 9.23TKK587 pKa = 10.53QLALTDD593 pKa = 4.07RR594 pKa = 11.84WFHH597 pKa = 5.37WEE599 pKa = 3.52WVIVRR604 pKa = 11.84KK605 pKa = 9.66FDD607 pKa = 4.3LPIIRR612 pKa = 11.84QFLVDD617 pKa = 4.46LRR619 pKa = 11.84KK620 pKa = 10.07LGYY623 pKa = 10.6DD624 pKa = 3.25KK625 pKa = 11.07AISKK629 pKa = 9.99NMSYY633 pKa = 10.02IGQVIQRR640 pKa = 11.84LPTLANDD647 pKa = 3.29YY648 pKa = 11.25AKK650 pKa = 10.11FVKK653 pKa = 10.06RR654 pKa = 11.84IYY656 pKa = 9.85MKK658 pKa = 10.77RR659 pKa = 11.84LPKK662 pKa = 10.02KK663 pKa = 9.99VNGAYY668 pKa = 9.6KK669 pKa = 10.16VFRR672 pKa = 11.84DD673 pKa = 3.62SLHH676 pKa = 4.95YY677 pKa = 9.55TINTKK682 pKa = 9.53VEE684 pKa = 3.82PHH686 pKa = 6.2TEE688 pKa = 3.77LGKK691 pKa = 9.67ATVVALNYY699 pKa = 10.05AVRR702 pKa = 11.84PTLPGNTCPYY712 pKa = 10.45DD713 pKa = 3.08SWSPFDD719 pKa = 3.85GDD721 pKa = 3.1VRR723 pKa = 11.84LIVV726 pKa = 3.42
Molecular weight: 83.79 kDa
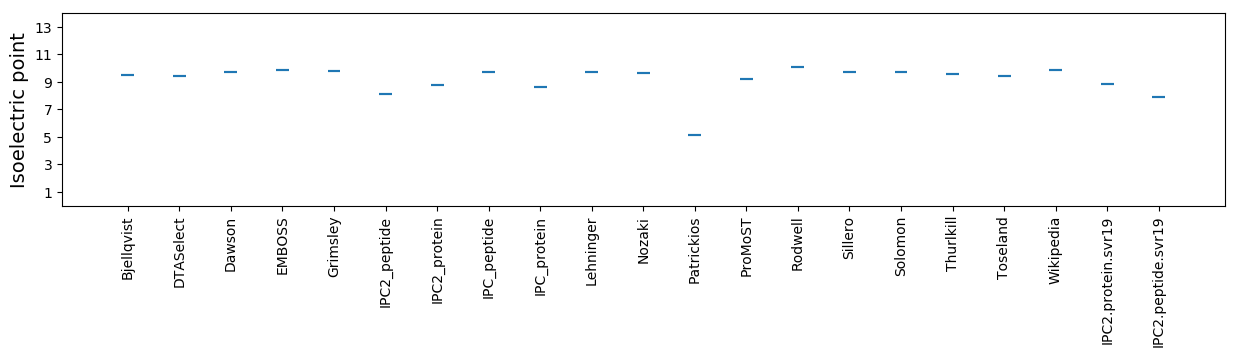
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIT8|A0A1L3KIT8_9VIRU RNA-dependent RNA polymerase OS=Wenzhou narna-like virus 7 OX=1923582 PE=4 SV=1
MM1 pKa = 7.44DD2 pKa = 4.22QEE4 pKa = 4.9FPCEE8 pKa = 3.84RR9 pKa = 11.84KK10 pKa = 9.17EE11 pKa = 4.64ALLDD15 pKa = 4.34HH16 pKa = 7.06INMKK20 pKa = 10.21IMSKK24 pKa = 10.14LRR26 pKa = 11.84NEE28 pKa = 4.04ARR30 pKa = 11.84HH31 pKa = 5.0KK32 pKa = 10.05RR33 pKa = 11.84KK34 pKa = 9.56RR35 pKa = 11.84AIGFAYY41 pKa = 10.38SIQNLKK47 pKa = 10.55YY48 pKa = 10.73LITFEE53 pKa = 4.57PKK55 pKa = 10.52DD56 pKa = 4.07LIKK59 pKa = 10.4STLEE63 pKa = 3.44KK64 pKa = 10.66HH65 pKa = 6.19RR66 pKa = 11.84ADD68 pKa = 3.24LSKK71 pKa = 10.63PIGGLDD77 pKa = 3.61PQIKK81 pKa = 9.25EE82 pKa = 4.11AYY84 pKa = 10.13DD85 pKa = 4.14FILQKK90 pKa = 10.93FMDD93 pKa = 5.75FIASNASEE101 pKa = 3.69IDD103 pKa = 3.73VFPEE107 pKa = 3.82PTEE110 pKa = 4.22SRR112 pKa = 11.84CHH114 pKa = 6.0SKK116 pKa = 9.07TGKK119 pKa = 9.6SFKK122 pKa = 9.87EE123 pKa = 3.99AFMKK127 pKa = 9.69ATSEE131 pKa = 4.09HH132 pKa = 6.39TSLCSNYY139 pKa = 9.63HH140 pKa = 5.38IRR142 pKa = 11.84VGDD145 pKa = 3.63RR146 pKa = 11.84TLCVPDD152 pKa = 3.39TNYY155 pKa = 11.08NKK157 pKa = 10.48DD158 pKa = 3.12FNFAHH163 pKa = 7.25GDD165 pKa = 3.61CLLNNTVKK173 pKa = 10.6AVALVEE179 pKa = 4.01PLKK182 pKa = 11.06VRR184 pKa = 11.84VITKK188 pKa = 8.8EE189 pKa = 3.81AAWTHH194 pKa = 4.87MLKK197 pKa = 10.47PIQKK201 pKa = 9.39LMHH204 pKa = 6.45SALTHH209 pKa = 6.39HH210 pKa = 7.36DD211 pKa = 3.52FGEE214 pKa = 4.47IFCLTRR220 pKa = 11.84GKK222 pKa = 9.83PYY224 pKa = 10.83EE225 pKa = 4.15NVSLGTLSDD234 pKa = 3.84SEE236 pKa = 5.14FYY238 pKa = 11.22LSGDD242 pKa = 3.89FSAATDD248 pKa = 3.89NIHH251 pKa = 6.53PHH253 pKa = 6.15FLNEE257 pKa = 3.43FRR259 pKa = 11.84DD260 pKa = 3.68RR261 pKa = 11.84MRR263 pKa = 11.84VILKK267 pKa = 7.87DD268 pKa = 3.53HH269 pKa = 6.22PMIWHH274 pKa = 7.33ALNSEE279 pKa = 4.02ARR281 pKa = 11.84NHH283 pKa = 6.25IIEE286 pKa = 4.55YY287 pKa = 9.52PLGIPPVEE295 pKa = 4.13QMRR298 pKa = 11.84GQLMGSLLSFPILCLSNLAIVLASCRR324 pKa = 11.84EE325 pKa = 4.22HH326 pKa = 7.39RR327 pKa = 11.84DD328 pKa = 3.16HH329 pKa = 7.22TIDD332 pKa = 4.83RR333 pKa = 11.84VIQSASSRR341 pKa = 11.84TGYY344 pKa = 10.55KK345 pKa = 10.2PIIEE349 pKa = 4.26KK350 pKa = 10.8VKK352 pKa = 9.5ITISEE357 pKa = 4.12NWSSTLEE364 pKa = 3.98RR365 pKa = 11.84LKK367 pKa = 10.91SSCLINGDD375 pKa = 3.89DD376 pKa = 4.51LAIRR380 pKa = 11.84TNDD383 pKa = 3.16QFLSKK388 pKa = 10.14WKK390 pKa = 10.31VLSPNAGFTLSIGKK404 pKa = 8.82NFIDD408 pKa = 4.24KK409 pKa = 10.46RR410 pKa = 11.84FYY412 pKa = 10.55TINSKK417 pKa = 10.77LFIDD421 pKa = 4.46DD422 pKa = 5.24KK423 pKa = 11.21IFSTHH428 pKa = 6.0KK429 pKa = 10.06FKK431 pKa = 11.23CSMSNKK437 pKa = 9.61GDD439 pKa = 3.8LASLGAEE446 pKa = 3.79YY447 pKa = 9.7PRR449 pKa = 11.84FFKK452 pKa = 10.85TEE454 pKa = 3.77GVQQEE459 pKa = 4.66VVDD462 pKa = 3.43KK463 pKa = 10.76HH464 pKa = 5.65NRR466 pKa = 11.84TIRR469 pKa = 11.84SMFIRR474 pKa = 11.84DD475 pKa = 3.17HH476 pKa = 6.76HH477 pKa = 6.57KK478 pKa = 10.86RR479 pKa = 11.84LMKK482 pKa = 10.39DD483 pKa = 3.19PRR485 pKa = 11.84SLEE488 pKa = 4.19VPRR491 pKa = 11.84HH492 pKa = 4.96LGGLGFGNLIPCNIMQKK509 pKa = 8.64LTHH512 pKa = 7.14LIYY515 pKa = 10.37SYY517 pKa = 11.07KK518 pKa = 8.81ITKK521 pKa = 9.52SPKK524 pKa = 9.69NFTNLLSRR532 pKa = 11.84FGLEE536 pKa = 3.07RR537 pKa = 11.84VYY539 pKa = 10.98VPRR542 pKa = 11.84GTCHH546 pKa = 7.15RR547 pKa = 11.84LKK549 pKa = 10.9LCDD552 pKa = 3.4YY553 pKa = 8.98TSYY556 pKa = 10.78IGLQFDD562 pKa = 3.85EE563 pKa = 4.71PTRR566 pKa = 11.84DD567 pKa = 3.17ISPSEE572 pKa = 3.79LPVRR576 pKa = 11.84IRR578 pKa = 11.84QYY580 pKa = 10.91IKK582 pKa = 10.63LVKK585 pKa = 9.23TKK587 pKa = 10.53QLALTDD593 pKa = 4.07RR594 pKa = 11.84WFHH597 pKa = 5.37WEE599 pKa = 3.52WVIVRR604 pKa = 11.84KK605 pKa = 9.66FDD607 pKa = 4.3LPIIRR612 pKa = 11.84QFLVDD617 pKa = 4.46LRR619 pKa = 11.84KK620 pKa = 10.07LGYY623 pKa = 10.6DD624 pKa = 3.25KK625 pKa = 11.07AISKK629 pKa = 9.99NMSYY633 pKa = 10.02IGQVIQRR640 pKa = 11.84LPTLANDD647 pKa = 3.29YY648 pKa = 11.25AKK650 pKa = 10.11FVKK653 pKa = 10.06RR654 pKa = 11.84IYY656 pKa = 9.85MKK658 pKa = 10.77RR659 pKa = 11.84LPKK662 pKa = 10.02KK663 pKa = 9.99VNGAYY668 pKa = 9.6KK669 pKa = 10.16VFRR672 pKa = 11.84DD673 pKa = 3.62SLHH676 pKa = 4.95YY677 pKa = 9.55TINTKK682 pKa = 9.53VEE684 pKa = 3.82PHH686 pKa = 6.2TEE688 pKa = 3.77LGKK691 pKa = 9.67ATVVALNYY699 pKa = 10.05AVRR702 pKa = 11.84PTLPGNTCPYY712 pKa = 10.45DD713 pKa = 3.08SWSPFDD719 pKa = 3.85GDD721 pKa = 3.1VRR723 pKa = 11.84LIVV726 pKa = 3.42
MM1 pKa = 7.44DD2 pKa = 4.22QEE4 pKa = 4.9FPCEE8 pKa = 3.84RR9 pKa = 11.84KK10 pKa = 9.17EE11 pKa = 4.64ALLDD15 pKa = 4.34HH16 pKa = 7.06INMKK20 pKa = 10.21IMSKK24 pKa = 10.14LRR26 pKa = 11.84NEE28 pKa = 4.04ARR30 pKa = 11.84HH31 pKa = 5.0KK32 pKa = 10.05RR33 pKa = 11.84KK34 pKa = 9.56RR35 pKa = 11.84AIGFAYY41 pKa = 10.38SIQNLKK47 pKa = 10.55YY48 pKa = 10.73LITFEE53 pKa = 4.57PKK55 pKa = 10.52DD56 pKa = 4.07LIKK59 pKa = 10.4STLEE63 pKa = 3.44KK64 pKa = 10.66HH65 pKa = 6.19RR66 pKa = 11.84ADD68 pKa = 3.24LSKK71 pKa = 10.63PIGGLDD77 pKa = 3.61PQIKK81 pKa = 9.25EE82 pKa = 4.11AYY84 pKa = 10.13DD85 pKa = 4.14FILQKK90 pKa = 10.93FMDD93 pKa = 5.75FIASNASEE101 pKa = 3.69IDD103 pKa = 3.73VFPEE107 pKa = 3.82PTEE110 pKa = 4.22SRR112 pKa = 11.84CHH114 pKa = 6.0SKK116 pKa = 9.07TGKK119 pKa = 9.6SFKK122 pKa = 9.87EE123 pKa = 3.99AFMKK127 pKa = 9.69ATSEE131 pKa = 4.09HH132 pKa = 6.39TSLCSNYY139 pKa = 9.63HH140 pKa = 5.38IRR142 pKa = 11.84VGDD145 pKa = 3.63RR146 pKa = 11.84TLCVPDD152 pKa = 3.39TNYY155 pKa = 11.08NKK157 pKa = 10.48DD158 pKa = 3.12FNFAHH163 pKa = 7.25GDD165 pKa = 3.61CLLNNTVKK173 pKa = 10.6AVALVEE179 pKa = 4.01PLKK182 pKa = 11.06VRR184 pKa = 11.84VITKK188 pKa = 8.8EE189 pKa = 3.81AAWTHH194 pKa = 4.87MLKK197 pKa = 10.47PIQKK201 pKa = 9.39LMHH204 pKa = 6.45SALTHH209 pKa = 6.39HH210 pKa = 7.36DD211 pKa = 3.52FGEE214 pKa = 4.47IFCLTRR220 pKa = 11.84GKK222 pKa = 9.83PYY224 pKa = 10.83EE225 pKa = 4.15NVSLGTLSDD234 pKa = 3.84SEE236 pKa = 5.14FYY238 pKa = 11.22LSGDD242 pKa = 3.89FSAATDD248 pKa = 3.89NIHH251 pKa = 6.53PHH253 pKa = 6.15FLNEE257 pKa = 3.43FRR259 pKa = 11.84DD260 pKa = 3.68RR261 pKa = 11.84MRR263 pKa = 11.84VILKK267 pKa = 7.87DD268 pKa = 3.53HH269 pKa = 6.22PMIWHH274 pKa = 7.33ALNSEE279 pKa = 4.02ARR281 pKa = 11.84NHH283 pKa = 6.25IIEE286 pKa = 4.55YY287 pKa = 9.52PLGIPPVEE295 pKa = 4.13QMRR298 pKa = 11.84GQLMGSLLSFPILCLSNLAIVLASCRR324 pKa = 11.84EE325 pKa = 4.22HH326 pKa = 7.39RR327 pKa = 11.84DD328 pKa = 3.16HH329 pKa = 7.22TIDD332 pKa = 4.83RR333 pKa = 11.84VIQSASSRR341 pKa = 11.84TGYY344 pKa = 10.55KK345 pKa = 10.2PIIEE349 pKa = 4.26KK350 pKa = 10.8VKK352 pKa = 9.5ITISEE357 pKa = 4.12NWSSTLEE364 pKa = 3.98RR365 pKa = 11.84LKK367 pKa = 10.91SSCLINGDD375 pKa = 3.89DD376 pKa = 4.51LAIRR380 pKa = 11.84TNDD383 pKa = 3.16QFLSKK388 pKa = 10.14WKK390 pKa = 10.31VLSPNAGFTLSIGKK404 pKa = 8.82NFIDD408 pKa = 4.24KK409 pKa = 10.46RR410 pKa = 11.84FYY412 pKa = 10.55TINSKK417 pKa = 10.77LFIDD421 pKa = 4.46DD422 pKa = 5.24KK423 pKa = 11.21IFSTHH428 pKa = 6.0KK429 pKa = 10.06FKK431 pKa = 11.23CSMSNKK437 pKa = 9.61GDD439 pKa = 3.8LASLGAEE446 pKa = 3.79YY447 pKa = 9.7PRR449 pKa = 11.84FFKK452 pKa = 10.85TEE454 pKa = 3.77GVQQEE459 pKa = 4.66VVDD462 pKa = 3.43KK463 pKa = 10.76HH464 pKa = 5.65NRR466 pKa = 11.84TIRR469 pKa = 11.84SMFIRR474 pKa = 11.84DD475 pKa = 3.17HH476 pKa = 6.76HH477 pKa = 6.57KK478 pKa = 10.86RR479 pKa = 11.84LMKK482 pKa = 10.39DD483 pKa = 3.19PRR485 pKa = 11.84SLEE488 pKa = 4.19VPRR491 pKa = 11.84HH492 pKa = 4.96LGGLGFGNLIPCNIMQKK509 pKa = 8.64LTHH512 pKa = 7.14LIYY515 pKa = 10.37SYY517 pKa = 11.07KK518 pKa = 8.81ITKK521 pKa = 9.52SPKK524 pKa = 9.69NFTNLLSRR532 pKa = 11.84FGLEE536 pKa = 3.07RR537 pKa = 11.84VYY539 pKa = 10.98VPRR542 pKa = 11.84GTCHH546 pKa = 7.15RR547 pKa = 11.84LKK549 pKa = 10.9LCDD552 pKa = 3.4YY553 pKa = 8.98TSYY556 pKa = 10.78IGLQFDD562 pKa = 3.85EE563 pKa = 4.71PTRR566 pKa = 11.84DD567 pKa = 3.17ISPSEE572 pKa = 3.79LPVRR576 pKa = 11.84IRR578 pKa = 11.84QYY580 pKa = 10.91IKK582 pKa = 10.63LVKK585 pKa = 9.23TKK587 pKa = 10.53QLALTDD593 pKa = 4.07RR594 pKa = 11.84WFHH597 pKa = 5.37WEE599 pKa = 3.52WVIVRR604 pKa = 11.84KK605 pKa = 9.66FDD607 pKa = 4.3LPIIRR612 pKa = 11.84QFLVDD617 pKa = 4.46LRR619 pKa = 11.84KK620 pKa = 10.07LGYY623 pKa = 10.6DD624 pKa = 3.25KK625 pKa = 11.07AISKK629 pKa = 9.99NMSYY633 pKa = 10.02IGQVIQRR640 pKa = 11.84LPTLANDD647 pKa = 3.29YY648 pKa = 11.25AKK650 pKa = 10.11FVKK653 pKa = 10.06RR654 pKa = 11.84IYY656 pKa = 9.85MKK658 pKa = 10.77RR659 pKa = 11.84LPKK662 pKa = 10.02KK663 pKa = 9.99VNGAYY668 pKa = 9.6KK669 pKa = 10.16VFRR672 pKa = 11.84DD673 pKa = 3.62SLHH676 pKa = 4.95YY677 pKa = 9.55TINTKK682 pKa = 9.53VEE684 pKa = 3.82PHH686 pKa = 6.2TEE688 pKa = 3.77LGKK691 pKa = 9.67ATVVALNYY699 pKa = 10.05AVRR702 pKa = 11.84PTLPGNTCPYY712 pKa = 10.45DD713 pKa = 3.08SWSPFDD719 pKa = 3.85GDD721 pKa = 3.1VRR723 pKa = 11.84LIVV726 pKa = 3.42
Molecular weight: 83.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
726 |
726 |
726 |
726.0 |
83.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.821 ± 0.0 | 1.928 ± 0.0 |
5.785 ± 0.0 | 4.821 ± 0.0 |
5.234 ± 0.0 | 4.545 ± 0.0 |
3.857 ± 0.0 | 7.576 ± 0.0 |
8.402 ± 0.0 | 10.055 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.342 ± 0.0 | 4.683 ± 0.0 |
4.821 ± 0.0 | 2.479 ± 0.0 |
6.474 ± 0.0 | 7.163 ± 0.0 |
5.51 ± 0.0 | 4.959 ± 0.0 |
1.102 ± 0.0 | 3.444 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
