
Tobacco mosaic virus (strain vulgare) (TMV) (Tobacco mosaic virus (strain U1))
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus; Tobacco mosaic virus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
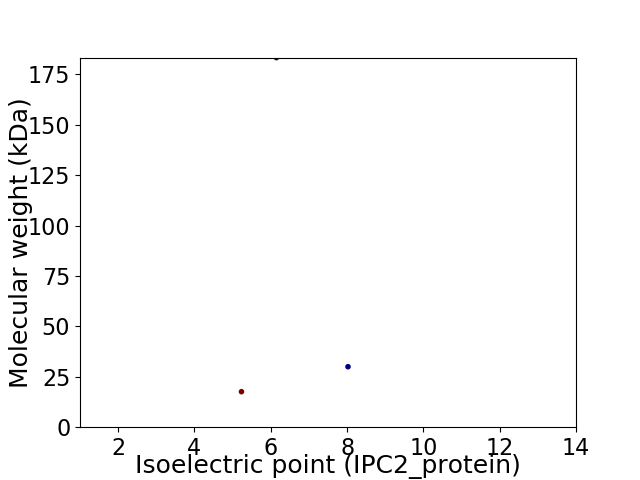
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
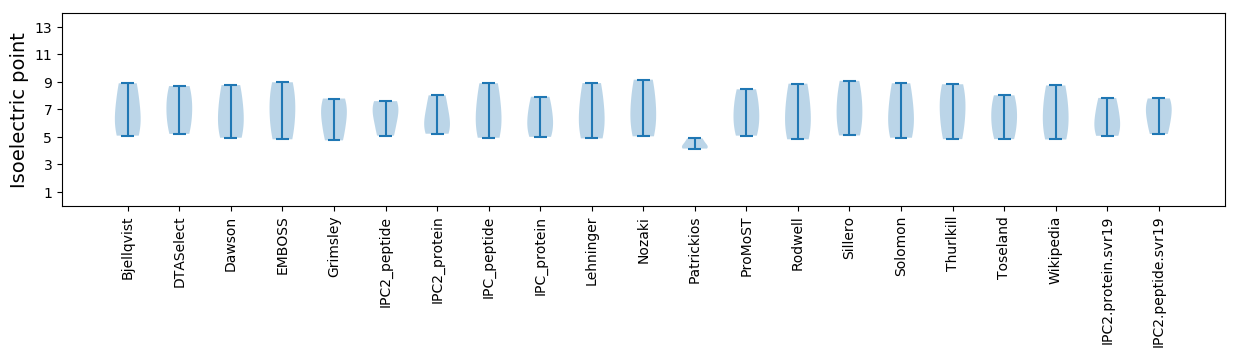
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P69687|CAPSD_TMV Capsid protein OS=Tobacco mosaic virus (strain vulgare) OX=12243 GN=CP PE=1 SV=2
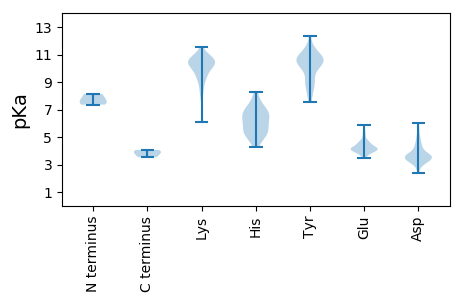
MM1 pKa = 7.62SYY3 pKa = 10.89SITTPSQFVFLSSAWADD20 pKa = 3.75PIEE23 pKa = 5.36LINLCTNALGNQFQTQQARR42 pKa = 11.84TVVQRR47 pKa = 11.84QFSEE51 pKa = 3.99VWKK54 pKa = 9.58PSPQVTVRR62 pKa = 11.84FPDD65 pKa = 3.15SDD67 pKa = 3.76FKK69 pKa = 11.2VYY71 pKa = 10.38RR72 pKa = 11.84YY73 pKa = 10.06NAVLDD78 pKa = 4.18PLVTALLGAFDD89 pKa = 3.55TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.22VEE98 pKa = 4.09NQANPTTAEE107 pKa = 4.03TLDD110 pKa = 3.11ATRR113 pKa = 11.84RR114 pKa = 11.84VDD116 pKa = 3.5DD117 pKa = 3.56ATVAIRR123 pKa = 11.84SAINNLIVEE132 pKa = 5.3LIRR135 pKa = 11.84GTGSYY140 pKa = 10.17NRR142 pKa = 11.84SSFEE146 pKa = 4.06SSSGLVWTSGPATT159 pKa = 3.54
MM1 pKa = 7.62SYY3 pKa = 10.89SITTPSQFVFLSSAWADD20 pKa = 3.75PIEE23 pKa = 5.36LINLCTNALGNQFQTQQARR42 pKa = 11.84TVVQRR47 pKa = 11.84QFSEE51 pKa = 3.99VWKK54 pKa = 9.58PSPQVTVRR62 pKa = 11.84FPDD65 pKa = 3.15SDD67 pKa = 3.76FKK69 pKa = 11.2VYY71 pKa = 10.38RR72 pKa = 11.84YY73 pKa = 10.06NAVLDD78 pKa = 4.18PLVTALLGAFDD89 pKa = 3.55TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.22VEE98 pKa = 4.09NQANPTTAEE107 pKa = 4.03TLDD110 pKa = 3.11ATRR113 pKa = 11.84RR114 pKa = 11.84VDD116 pKa = 3.5DD117 pKa = 3.56ATVAIRR123 pKa = 11.84SAINNLIVEE132 pKa = 5.3LIRR135 pKa = 11.84GTGSYY140 pKa = 10.17NRR142 pKa = 11.84SSFEE146 pKa = 4.06SSSGLVWTSGPATT159 pKa = 3.54
Molecular weight: 17.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03586|RDRP_TMV Replicase large subunit OS=Tobacco mosaic virus (strain vulgare) OX=12243 PE=1 SV=2
MM1 pKa = 7.36ALVVKK6 pKa = 10.26GKK8 pKa = 10.77VNINEE13 pKa = 4.75FIDD16 pKa = 3.67LTKK19 pKa = 9.99MEE21 pKa = 5.53KK22 pKa = 9.76ILPSMFTPVKK32 pKa = 10.48SVMCSKK38 pKa = 10.25VDD40 pKa = 3.36KK41 pKa = 11.19IMVHH45 pKa = 5.51EE46 pKa = 4.48NEE48 pKa = 4.63SLSEE52 pKa = 4.11VNLLKK57 pKa = 10.53GVKK60 pKa = 9.92LIDD63 pKa = 3.41SGYY66 pKa = 8.75VCLAGLVVTGEE77 pKa = 3.84WNLPDD82 pKa = 3.53NCRR85 pKa = 11.84GGVSVCLVDD94 pKa = 5.03KK95 pKa = 10.99RR96 pKa = 11.84MEE98 pKa = 4.33RR99 pKa = 11.84ADD101 pKa = 3.63EE102 pKa = 4.26ATLGSYY108 pKa = 7.68YY109 pKa = 10.0TAAAKK114 pKa = 10.21KK115 pKa = 8.92RR116 pKa = 11.84FQFKK120 pKa = 9.97VVPNYY125 pKa = 10.93AITTQDD131 pKa = 2.4AMKK134 pKa = 10.59NVWQVLVNIRR144 pKa = 11.84NVKK147 pKa = 9.22MSAGFCPLSLEE158 pKa = 4.22FVSVCIVYY166 pKa = 10.53RR167 pKa = 11.84NNIKK171 pKa = 10.59LGLRR175 pKa = 11.84EE176 pKa = 4.48KK177 pKa = 9.44ITNVRR182 pKa = 11.84DD183 pKa = 3.72GGPMEE188 pKa = 4.18LTEE191 pKa = 4.11EE192 pKa = 4.32VVDD195 pKa = 4.43EE196 pKa = 4.56FMEE199 pKa = 4.33DD200 pKa = 3.01VPMSIRR206 pKa = 11.84LAKK209 pKa = 10.0FRR211 pKa = 11.84SRR213 pKa = 11.84TGKK216 pKa = 10.45KK217 pKa = 9.53SDD219 pKa = 3.21VRR221 pKa = 11.84KK222 pKa = 10.33GKK224 pKa = 10.15NSSNDD229 pKa = 2.77RR230 pKa = 11.84SVPNKK235 pKa = 10.1NYY237 pKa = 10.64RR238 pKa = 11.84NVKK241 pKa = 10.28DD242 pKa = 3.95FGGMSFKK249 pKa = 10.82KK250 pKa = 10.59NNLIDD255 pKa = 5.08DD256 pKa = 4.44DD257 pKa = 4.9SEE259 pKa = 4.37ATVAEE264 pKa = 4.3SDD266 pKa = 3.89SFF268 pKa = 3.87
MM1 pKa = 7.36ALVVKK6 pKa = 10.26GKK8 pKa = 10.77VNINEE13 pKa = 4.75FIDD16 pKa = 3.67LTKK19 pKa = 9.99MEE21 pKa = 5.53KK22 pKa = 9.76ILPSMFTPVKK32 pKa = 10.48SVMCSKK38 pKa = 10.25VDD40 pKa = 3.36KK41 pKa = 11.19IMVHH45 pKa = 5.51EE46 pKa = 4.48NEE48 pKa = 4.63SLSEE52 pKa = 4.11VNLLKK57 pKa = 10.53GVKK60 pKa = 9.92LIDD63 pKa = 3.41SGYY66 pKa = 8.75VCLAGLVVTGEE77 pKa = 3.84WNLPDD82 pKa = 3.53NCRR85 pKa = 11.84GGVSVCLVDD94 pKa = 5.03KK95 pKa = 10.99RR96 pKa = 11.84MEE98 pKa = 4.33RR99 pKa = 11.84ADD101 pKa = 3.63EE102 pKa = 4.26ATLGSYY108 pKa = 7.68YY109 pKa = 10.0TAAAKK114 pKa = 10.21KK115 pKa = 8.92RR116 pKa = 11.84FQFKK120 pKa = 9.97VVPNYY125 pKa = 10.93AITTQDD131 pKa = 2.4AMKK134 pKa = 10.59NVWQVLVNIRR144 pKa = 11.84NVKK147 pKa = 9.22MSAGFCPLSLEE158 pKa = 4.22FVSVCIVYY166 pKa = 10.53RR167 pKa = 11.84NNIKK171 pKa = 10.59LGLRR175 pKa = 11.84EE176 pKa = 4.48KK177 pKa = 9.44ITNVRR182 pKa = 11.84DD183 pKa = 3.72GGPMEE188 pKa = 4.18LTEE191 pKa = 4.11EE192 pKa = 4.32VVDD195 pKa = 4.43EE196 pKa = 4.56FMEE199 pKa = 4.33DD200 pKa = 3.01VPMSIRR206 pKa = 11.84LAKK209 pKa = 10.0FRR211 pKa = 11.84SRR213 pKa = 11.84TGKK216 pKa = 10.45KK217 pKa = 9.53SDD219 pKa = 3.21VRR221 pKa = 11.84KK222 pKa = 10.33GKK224 pKa = 10.15NSSNDD229 pKa = 2.77RR230 pKa = 11.84SVPNKK235 pKa = 10.1NYY237 pKa = 10.64RR238 pKa = 11.84NVKK241 pKa = 10.28DD242 pKa = 3.95FGGMSFKK249 pKa = 10.82KK250 pKa = 10.59NNLIDD255 pKa = 5.08DD256 pKa = 4.44DD257 pKa = 4.9SEE259 pKa = 4.37ATVAEE264 pKa = 4.3SDD266 pKa = 3.89SFF268 pKa = 3.87
Molecular weight: 30.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2043 |
159 |
1616 |
681.0 |
76.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.755 ± 0.582 | 1.811 ± 0.248 |
6.314 ± 0.301 | 5.874 ± 0.308 |
4.748 ± 0.189 | 4.503 ± 0.428 |
2.056 ± 0.897 | 4.993 ± 0.168 |
6.559 ± 1.19 | 9.3 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.541 | 4.405 ± 1.282 |
3.377 ± 0.351 | 3.279 ± 0.664 |
5.384 ± 0.344 | 8.37 ± 0.355 |
5.776 ± 0.9 | 8.762 ± 1.05 |
0.979 ± 0.192 | 3.769 ± 0.697 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
