
Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; Deinococcus radiodurans
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
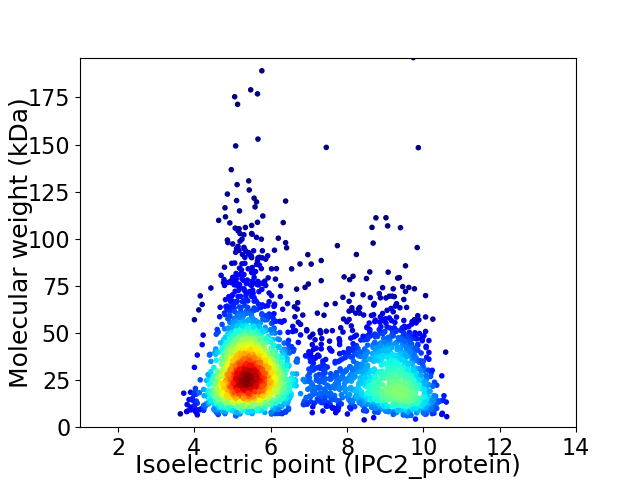
Virtual 2D-PAGE plot for 3085 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9RZT0|Q9RZT0_DEIRA Arylesterase/monoxygenase OS=Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) OX=243230 GN=DR_B0033 PE=3 SV=1
MM1 pKa = 7.85DD2 pKa = 5.29LSEE5 pKa = 4.12LAHH8 pKa = 6.9RR9 pKa = 11.84ITYY12 pKa = 9.63RR13 pKa = 11.84AYY15 pKa = 10.05EE16 pKa = 4.16LDD18 pKa = 4.33GDD20 pKa = 4.81DD21 pKa = 5.1LDD23 pKa = 5.42SLAGLCGLMSWHH35 pKa = 6.33TLIAPLTFQEE45 pKa = 5.14FGTEE49 pKa = 3.9DD50 pKa = 3.93GRR52 pKa = 11.84TLLCAADD59 pKa = 4.85EE60 pKa = 4.47SGLWITLTDD69 pKa = 3.98GAAGVPTSPDD79 pKa = 3.11TFQLSLAEE87 pKa = 4.81DD88 pKa = 4.08LLSEE92 pKa = 4.17PVYY95 pKa = 10.53TLDD98 pKa = 3.25VVNGHH103 pKa = 5.84VVQTAPGLNN112 pKa = 3.47
MM1 pKa = 7.85DD2 pKa = 5.29LSEE5 pKa = 4.12LAHH8 pKa = 6.9RR9 pKa = 11.84ITYY12 pKa = 9.63RR13 pKa = 11.84AYY15 pKa = 10.05EE16 pKa = 4.16LDD18 pKa = 4.33GDD20 pKa = 4.81DD21 pKa = 5.1LDD23 pKa = 5.42SLAGLCGLMSWHH35 pKa = 6.33TLIAPLTFQEE45 pKa = 5.14FGTEE49 pKa = 3.9DD50 pKa = 3.93GRR52 pKa = 11.84TLLCAADD59 pKa = 4.85EE60 pKa = 4.47SGLWITLTDD69 pKa = 3.98GAAGVPTSPDD79 pKa = 3.11TFQLSLAEE87 pKa = 4.81DD88 pKa = 4.08LLSEE92 pKa = 4.17PVYY95 pKa = 10.53TLDD98 pKa = 3.25VVNGHH103 pKa = 5.84VVQTAPGLNN112 pKa = 3.47
Molecular weight: 12.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9RWZ6|Q9RWZ6_DEIRA Uncharacterized protein OS=Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) OX=243230 GN=DR_0519 PE=4 SV=1
MM1 pKa = 7.58LCGGTRR7 pKa = 11.84GVVSKK12 pKa = 11.03AVGPFPGQGGEE23 pKa = 4.01QARR26 pKa = 11.84QRR28 pKa = 11.84GPQRR32 pKa = 11.84QGGSEE37 pKa = 4.33MVWEE41 pKa = 4.49TLPEE45 pKa = 4.18AASLFACSSPGAKK58 pKa = 9.73AGRR61 pKa = 11.84TARR64 pKa = 11.84QHH66 pKa = 4.98STLARR71 pKa = 11.84ALAAASPSDD80 pKa = 3.53RR81 pKa = 11.84TCTVLSQKK89 pKa = 10.49AQARR93 pKa = 11.84PAGQNFRR100 pKa = 11.84WGPTQGNVKK109 pKa = 9.35PRR111 pKa = 11.84KK112 pKa = 8.39RR113 pKa = 11.84NVRR116 pKa = 11.84NFRR119 pKa = 11.84LLYY122 pKa = 8.44GQAGAGIPVTANAISRR138 pKa = 11.84QRR140 pKa = 11.84GTIHH144 pKa = 6.49TLSAAPNHH152 pKa = 6.55RR153 pKa = 11.84RR154 pKa = 11.84FTMKK158 pKa = 9.45NTTQGFTLIEE168 pKa = 4.17LLIVIAIIGILAAVLIPNLLGARR191 pKa = 11.84AKK193 pKa = 10.99ANDD196 pKa = 3.96SATQSYY202 pKa = 9.98IRR204 pKa = 11.84NCYY207 pKa = 8.03TAVEE211 pKa = 4.5VARR214 pKa = 11.84GADD217 pKa = 3.33GRR219 pKa = 11.84LPAAGNCDD227 pKa = 3.02STANLGTNAVATPGAITAAGAYY249 pKa = 9.94SLDD252 pKa = 3.44GTGSKK257 pKa = 10.3FAVTATSVTNKK268 pKa = 8.3TFCFDD273 pKa = 3.43GAKK276 pKa = 9.61MGEE279 pKa = 4.29GTGATGACTFSTSTGTGTGTGTGTGTGTGTGTGSNN314 pKa = 3.8
MM1 pKa = 7.58LCGGTRR7 pKa = 11.84GVVSKK12 pKa = 11.03AVGPFPGQGGEE23 pKa = 4.01QARR26 pKa = 11.84QRR28 pKa = 11.84GPQRR32 pKa = 11.84QGGSEE37 pKa = 4.33MVWEE41 pKa = 4.49TLPEE45 pKa = 4.18AASLFACSSPGAKK58 pKa = 9.73AGRR61 pKa = 11.84TARR64 pKa = 11.84QHH66 pKa = 4.98STLARR71 pKa = 11.84ALAAASPSDD80 pKa = 3.53RR81 pKa = 11.84TCTVLSQKK89 pKa = 10.49AQARR93 pKa = 11.84PAGQNFRR100 pKa = 11.84WGPTQGNVKK109 pKa = 9.35PRR111 pKa = 11.84KK112 pKa = 8.39RR113 pKa = 11.84NVRR116 pKa = 11.84NFRR119 pKa = 11.84LLYY122 pKa = 8.44GQAGAGIPVTANAISRR138 pKa = 11.84QRR140 pKa = 11.84GTIHH144 pKa = 6.49TLSAAPNHH152 pKa = 6.55RR153 pKa = 11.84RR154 pKa = 11.84FTMKK158 pKa = 9.45NTTQGFTLIEE168 pKa = 4.17LLIVIAIIGILAAVLIPNLLGARR191 pKa = 11.84AKK193 pKa = 10.99ANDD196 pKa = 3.96SATQSYY202 pKa = 9.98IRR204 pKa = 11.84NCYY207 pKa = 8.03TAVEE211 pKa = 4.5VARR214 pKa = 11.84GADD217 pKa = 3.33GRR219 pKa = 11.84LPAAGNCDD227 pKa = 3.02STANLGTNAVATPGAITAAGAYY249 pKa = 9.94SLDD252 pKa = 3.44GTGSKK257 pKa = 10.3FAVTATSVTNKK268 pKa = 8.3TFCFDD273 pKa = 3.43GAKK276 pKa = 9.61MGEE279 pKa = 4.29GTGATGACTFSTSTGTGTGTGTGTGTGTGTGTGSNN314 pKa = 3.8
Molecular weight: 31.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
949308 |
37 |
1940 |
307.7 |
33.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.205 ± 0.064 | 0.668 ± 0.013 |
5.069 ± 0.035 | 5.731 ± 0.046 |
3.156 ± 0.027 | 9.198 ± 0.053 |
2.085 ± 0.025 | 3.284 ± 0.037 |
2.717 ± 0.041 | 11.65 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.891 ± 0.023 | 2.407 ± 0.027 |
6.049 ± 0.041 | 4.118 ± 0.035 |
7.375 ± 0.042 | 5.2 ± 0.031 |
5.815 ± 0.041 | 7.687 ± 0.035 |
1.388 ± 0.017 | 2.303 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
