
Stachybotrys chlorohalonata (strain IBT 40285)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Stachybotryaceae; Stachybotrys; Stachybotrys chlorohalonata
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
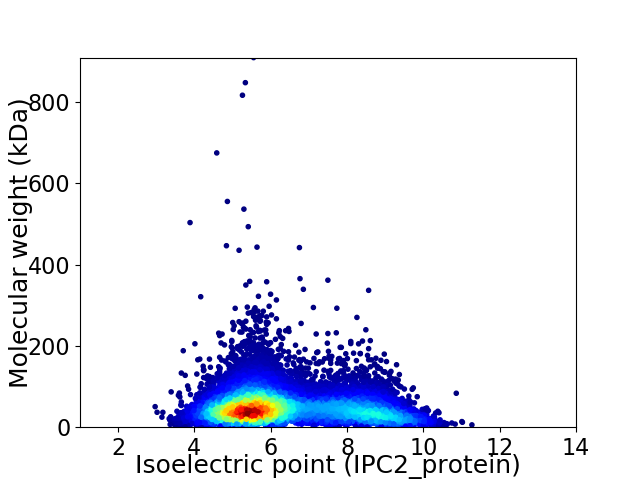
Virtual 2D-PAGE plot for 10676 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084QLP7|A0A084QLP7_STAC4 MHD domain-containing protein (Fragment) OS=Stachybotrys chlorohalonata (strain IBT 40285) OX=1283841 GN=S40285_02896 PE=4 SV=1
SS1 pKa = 6.49QLPNLLSLLSGLPGFPDD18 pKa = 3.15IATQTDD24 pKa = 4.09LPSIISALPNAPPALLPQLSTLFDD48 pKa = 4.46FLTQQPDD55 pKa = 3.62FPGILTGLPTPGLPLPTGLLSLAPPAASQLPNLLSLLSGLPGFPNIATQTDD106 pKa = 4.21LPSIISGLPAVPSGFLPQLPSLLDD130 pKa = 3.75FLTQQPDD137 pKa = 3.28LPGISTGLPTPGLPPTGLPSLAPSAASQLPNLLSLLSGLPGFPDD181 pKa = 3.06IATQIDD187 pKa = 4.51LPSIISGLPAVPSGLLPQLPTLFDD211 pKa = 3.94FLTRR215 pKa = 11.84QPNFPGILTGQPTPALPAFTGLPNIGSPVAGLPNFGFPSAPTLGGAQEE263 pKa = 4.29TEE265 pKa = 4.18LPKK268 pKa = 10.74VGSPTIPGWPNFGLPPFTGLPTLALPAASQLPNLLSILSGLPGFPNIATQTNLPSIISGLPNVPSGLLPQLPTLLDD344 pKa = 3.89FLTGQPNFPGILTAAPTFPGAPSFTGVPPVPGFPPLSGLPDD385 pKa = 3.76LPMPAMSQLPNLLNILSGLPGFPNIATQTNLPNIISGLPNVPSGLLPQLPTLLDD439 pKa = 3.89FLTGQPNFPGILTAAPTFPEE459 pKa = 4.74APSSTGLPNFGLPLASGLPPLTGLPGLPLSAISQLPNLLHH499 pKa = 6.15VLSGLPGFPNIATQTNLPSIVSGLPNAPSGLLPQLPTLLDD539 pKa = 3.68FLTRR543 pKa = 11.84QPNFPGVLTGTPTFPGLPTFPALPGFSGFPSAPGFPNIPGLPYY586 pKa = 10.76LPGLPTPPALPGPPGFPNIPGLPYY610 pKa = 10.76LPGLPTPPALPGFSGFPRR628 pKa = 11.84PPGFPNIPGLPYY640 pKa = 10.77LPDD643 pKa = 4.51LPTPPAFPSFPGFPGFPSPGGLPNKK668 pKa = 10.49LPGLPSASEE677 pKa = 4.42LPNQSGVPSIPGAPTFPSLPKK698 pKa = 10.31FSGPPTFPGLPNLPGLPNLPNPLPTALSQLPALLDD733 pKa = 3.78FLPGLPNFPEE743 pKa = 4.28VLTRR747 pKa = 11.84PSLPDD752 pKa = 3.89IISNAPNVPSGVLPQLPALLDD773 pKa = 3.81FLNGQPNFPGTPTSPPNLGNIFTGLPNLSPPAASQLPQLLNFLSEE818 pKa = 4.3LPSFPEE824 pKa = 3.97VLTQQALPSIISNAPNLPSGILPQLPEE851 pKa = 4.48LLNFLSGQPNFPAIPTGQPGLGSIFTGLPTLAPSAASQLPQLLSLLPSLPGFPNILTQPALPSIISNAPSVPSGVLPQLPEE932 pKa = 4.06LLNFLSQQPNFPGVLTGGLPGPGGIFNALPTLAPPAASQLPQLLNILPSLPGFPNILTQPALPSIISNAPNVPPGVLPEE1011 pKa = 4.33LPALLSFLSQQPNFPGILTGGPADD1035 pKa = 4.21LGSVFTGLPSLAPPAASQLPQLLSLLPSVPGFPNVLTASDD1075 pKa = 4.36LSSALAALPSLPPNLLTQLPTLLDD1099 pKa = 3.71FLTRR1103 pKa = 11.84QPNYY1107 pKa = 9.66PDD1109 pKa = 3.39IVTALPNFSLPAFVPAVPTSVPGMPQFPGIPTGGFQFPAAPTGVPGAPSFPGVPKK1164 pKa = 10.88APGKK1168 pKa = 9.64PSGVPAVPTGKK1179 pKa = 9.43PGTPGEE1185 pKa = 4.25PKK1187 pKa = 10.73LPVTNVLSGAPQFPGSPLFPSAPQLPEE1214 pKa = 4.54VPDD1217 pKa = 3.89KK1218 pKa = 10.37PTDD1221 pKa = 3.79PGAPQASDD1229 pKa = 3.64GADD1232 pKa = 3.57PLCPYY1237 pKa = 9.21PPWWEE1242 pKa = 3.87LQQPDD1247 pKa = 4.55DD1248 pKa = 5.27GVDD1251 pKa = 3.75EE1252 pKa = 4.38QCCCCDD1258 pKa = 2.48IWTGNIACRR1267 pKa = 11.84MLSGGCVCPAVEE1279 pKa = 4.72CAPGAPTVWPYY1290 pKa = 11.31GQDD1293 pKa = 2.89ATLPIATGIPGTDD1306 pKa = 3.55GYY1308 pKa = 11.05GIPAPP1313 pKa = 4.12
SS1 pKa = 6.49QLPNLLSLLSGLPGFPDD18 pKa = 3.15IATQTDD24 pKa = 4.09LPSIISALPNAPPALLPQLSTLFDD48 pKa = 4.46FLTQQPDD55 pKa = 3.62FPGILTGLPTPGLPLPTGLLSLAPPAASQLPNLLSLLSGLPGFPNIATQTDD106 pKa = 4.21LPSIISGLPAVPSGFLPQLPSLLDD130 pKa = 3.75FLTQQPDD137 pKa = 3.28LPGISTGLPTPGLPPTGLPSLAPSAASQLPNLLSLLSGLPGFPDD181 pKa = 3.06IATQIDD187 pKa = 4.51LPSIISGLPAVPSGLLPQLPTLFDD211 pKa = 3.94FLTRR215 pKa = 11.84QPNFPGILTGQPTPALPAFTGLPNIGSPVAGLPNFGFPSAPTLGGAQEE263 pKa = 4.29TEE265 pKa = 4.18LPKK268 pKa = 10.74VGSPTIPGWPNFGLPPFTGLPTLALPAASQLPNLLSILSGLPGFPNIATQTNLPSIISGLPNVPSGLLPQLPTLLDD344 pKa = 3.89FLTGQPNFPGILTAAPTFPGAPSFTGVPPVPGFPPLSGLPDD385 pKa = 3.76LPMPAMSQLPNLLNILSGLPGFPNIATQTNLPNIISGLPNVPSGLLPQLPTLLDD439 pKa = 3.89FLTGQPNFPGILTAAPTFPEE459 pKa = 4.74APSSTGLPNFGLPLASGLPPLTGLPGLPLSAISQLPNLLHH499 pKa = 6.15VLSGLPGFPNIATQTNLPSIVSGLPNAPSGLLPQLPTLLDD539 pKa = 3.68FLTRR543 pKa = 11.84QPNFPGVLTGTPTFPGLPTFPALPGFSGFPSAPGFPNIPGLPYY586 pKa = 10.76LPGLPTPPALPGPPGFPNIPGLPYY610 pKa = 10.76LPGLPTPPALPGFSGFPRR628 pKa = 11.84PPGFPNIPGLPYY640 pKa = 10.77LPDD643 pKa = 4.51LPTPPAFPSFPGFPGFPSPGGLPNKK668 pKa = 10.49LPGLPSASEE677 pKa = 4.42LPNQSGVPSIPGAPTFPSLPKK698 pKa = 10.31FSGPPTFPGLPNLPGLPNLPNPLPTALSQLPALLDD733 pKa = 3.78FLPGLPNFPEE743 pKa = 4.28VLTRR747 pKa = 11.84PSLPDD752 pKa = 3.89IISNAPNVPSGVLPQLPALLDD773 pKa = 3.81FLNGQPNFPGTPTSPPNLGNIFTGLPNLSPPAASQLPQLLNFLSEE818 pKa = 4.3LPSFPEE824 pKa = 3.97VLTQQALPSIISNAPNLPSGILPQLPEE851 pKa = 4.48LLNFLSGQPNFPAIPTGQPGLGSIFTGLPTLAPSAASQLPQLLSLLPSLPGFPNILTQPALPSIISNAPSVPSGVLPQLPEE932 pKa = 4.06LLNFLSQQPNFPGVLTGGLPGPGGIFNALPTLAPPAASQLPQLLNILPSLPGFPNILTQPALPSIISNAPNVPPGVLPEE1011 pKa = 4.33LPALLSFLSQQPNFPGILTGGPADD1035 pKa = 4.21LGSVFTGLPSLAPPAASQLPQLLSLLPSVPGFPNVLTASDD1075 pKa = 4.36LSSALAALPSLPPNLLTQLPTLLDD1099 pKa = 3.71FLTRR1103 pKa = 11.84QPNYY1107 pKa = 9.66PDD1109 pKa = 3.39IVTALPNFSLPAFVPAVPTSVPGMPQFPGIPTGGFQFPAAPTGVPGAPSFPGVPKK1164 pKa = 10.88APGKK1168 pKa = 9.64PSGVPAVPTGKK1179 pKa = 9.43PGTPGEE1185 pKa = 4.25PKK1187 pKa = 10.73LPVTNVLSGAPQFPGSPLFPSAPQLPEE1214 pKa = 4.54VPDD1217 pKa = 3.89KK1218 pKa = 10.37PTDD1221 pKa = 3.79PGAPQASDD1229 pKa = 3.64GADD1232 pKa = 3.57PLCPYY1237 pKa = 9.21PPWWEE1242 pKa = 3.87LQQPDD1247 pKa = 4.55DD1248 pKa = 5.27GVDD1251 pKa = 3.75EE1252 pKa = 4.38QCCCCDD1258 pKa = 2.48IWTGNIACRR1267 pKa = 11.84MLSGGCVCPAVEE1279 pKa = 4.72CAPGAPTVWPYY1290 pKa = 11.31GQDD1293 pKa = 2.89ATLPIATGIPGTDD1306 pKa = 3.55GYY1308 pKa = 11.05GIPAPP1313 pKa = 4.12
Molecular weight: 133.21 kDa
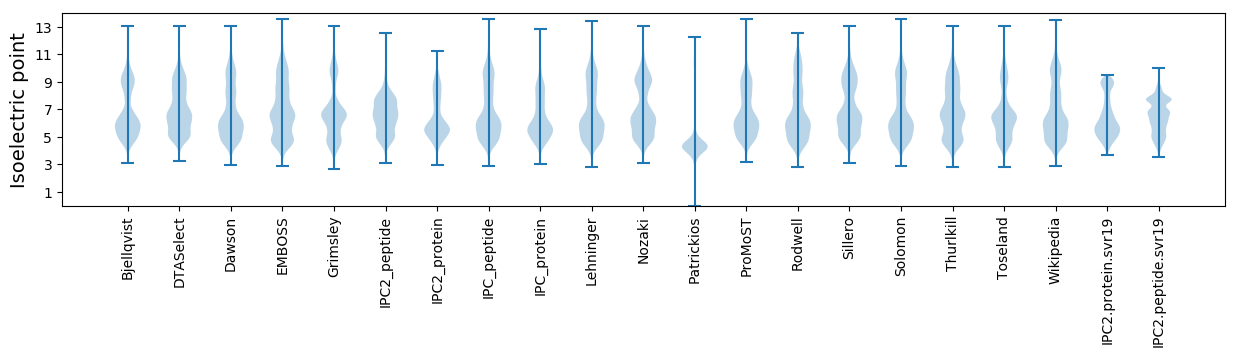
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A084QN67|A0A084QN67_STAC4 Tryptophanyl-tRNA synthetase OS=Stachybotrys chlorohalonata (strain IBT 40285) OX=1283841 GN=S40285_00439 PE=3 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84WNGTRR7 pKa = 11.84TSSTLSSPTLTTRR20 pKa = 11.84RR21 pKa = 11.84PASAGSASGRR31 pKa = 11.84SRR33 pKa = 11.84GRR35 pKa = 11.84PSPRR39 pKa = 11.84TRR41 pKa = 11.84STTNHH46 pKa = 5.44RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84TII52 pKa = 2.95
MM1 pKa = 7.74RR2 pKa = 11.84WNGTRR7 pKa = 11.84TSSTLSSPTLTTRR20 pKa = 11.84RR21 pKa = 11.84PASAGSASGRR31 pKa = 11.84SRR33 pKa = 11.84GRR35 pKa = 11.84PSPRR39 pKa = 11.84TRR41 pKa = 11.84STTNHH46 pKa = 5.44RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84TII52 pKa = 2.95
Molecular weight: 5.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5257513 |
8 |
8218 |
492.5 |
54.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.075 ± 0.025 | 1.215 ± 0.01 |
5.889 ± 0.018 | 6.064 ± 0.023 |
3.623 ± 0.015 | 6.948 ± 0.024 |
2.445 ± 0.011 | 4.677 ± 0.018 |
4.45 ± 0.021 | 8.937 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.009 | 3.536 ± 0.012 |
6.135 ± 0.026 | 4.057 ± 0.016 |
6.308 ± 0.023 | 8.057 ± 0.024 |
5.891 ± 0.023 | 6.276 ± 0.018 |
1.488 ± 0.009 | 2.694 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
