
Capybara microvirus Cap1_SP_45
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
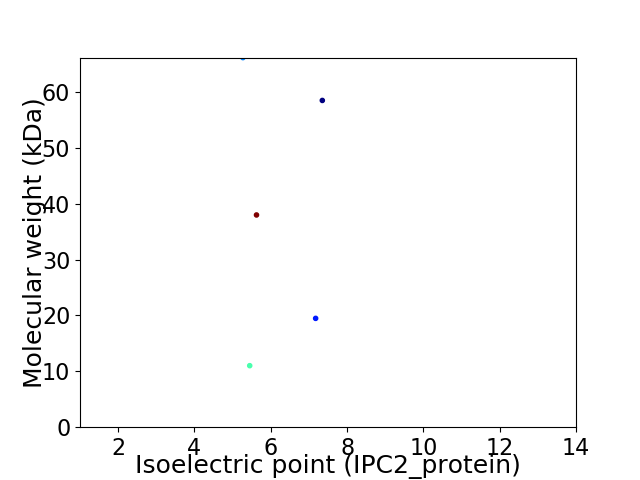
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
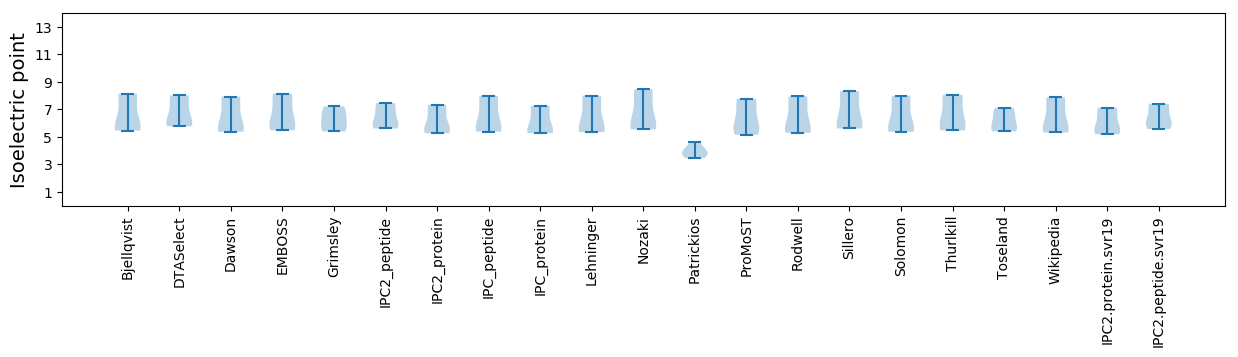
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVM4|A0A4V1FVM4_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_45 OX=2584778 PE=4 SV=1
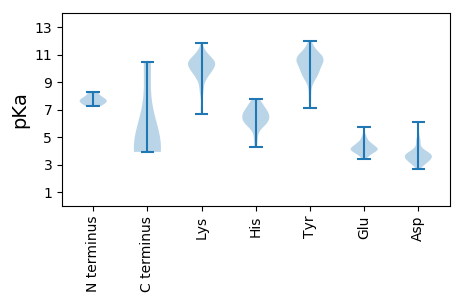
MM1 pKa = 7.61SNNNSQTINISKK13 pKa = 9.6ASKK16 pKa = 9.78YY17 pKa = 10.84SRR19 pKa = 11.84ISLDD23 pKa = 3.46EE24 pKa = 3.98AHH26 pKa = 6.81LTTLDD31 pKa = 3.88FGQIMPIYY39 pKa = 9.79SRR41 pKa = 11.84AIIPGDD47 pKa = 3.91DD48 pKa = 3.68LSIDD52 pKa = 3.7TNVFTRR58 pKa = 11.84TSPLVYY64 pKa = 9.73PSFVNAHH71 pKa = 5.7LTLVSVFVPYY81 pKa = 10.42YY82 pKa = 9.17QLSPFMEE89 pKa = 4.65PFFAGVPNIGSKK101 pKa = 8.27VVKK104 pKa = 10.52LPLLTNHH111 pKa = 7.33IIDD114 pKa = 4.81HH115 pKa = 6.85LFTDD119 pKa = 5.16LAQTLNIGKK128 pKa = 9.59IGQIGVNPDD137 pKa = 4.11IIWTTTDD144 pKa = 2.81NLKK147 pKa = 11.01YY148 pKa = 9.68GTQLNYY154 pKa = 10.4KK155 pKa = 8.78GRR157 pKa = 11.84LMLKK161 pKa = 9.54VLNSLGYY168 pKa = 9.38QVSKK172 pKa = 10.6NVSWKK177 pKa = 10.36SSVDD181 pKa = 3.41NEE183 pKa = 4.5YY184 pKa = 11.14KK185 pKa = 10.69NSPVTLNPLPFISYY199 pKa = 10.0IKK201 pKa = 10.43VYY203 pKa = 10.75KK204 pKa = 10.48DD205 pKa = 2.82WFSSNKK211 pKa = 9.88SFNTSYY217 pKa = 10.82VASILNRR224 pKa = 11.84IQTEE228 pKa = 3.99SDD230 pKa = 3.78FEE232 pKa = 4.1FTPQIINSLFDD243 pKa = 3.98YY244 pKa = 10.91LRR246 pKa = 11.84LLYY249 pKa = 9.73DD250 pKa = 3.18TDD252 pKa = 4.06YY253 pKa = 9.57FTTAWQYY260 pKa = 11.58NNYY263 pKa = 9.56PSDD266 pKa = 3.7SSKK269 pKa = 10.17TSTDD273 pKa = 3.06ILPDD277 pKa = 4.05PSWSGLYY284 pKa = 10.29DD285 pKa = 3.45GNRR288 pKa = 11.84FLINPNEE295 pKa = 3.98NLADD299 pKa = 3.61QSDD302 pKa = 4.27PEE304 pKa = 4.31FLGATTVRR312 pKa = 11.84LLSVLDD318 pKa = 3.54KK319 pKa = 10.72YY320 pKa = 10.89IRR322 pKa = 11.84RR323 pKa = 11.84NSYY326 pKa = 10.99AGTKK330 pKa = 9.28AAARR334 pKa = 11.84VLSRR338 pKa = 11.84FGIKK342 pKa = 8.54STEE345 pKa = 4.11FKK347 pKa = 10.69SQYY350 pKa = 10.89SDD352 pKa = 2.79ILNVDD357 pKa = 4.6SIRR360 pKa = 11.84LNIGDD365 pKa = 3.8VTSTAEE371 pKa = 4.11TTEE374 pKa = 4.2MPLGSYY380 pKa = 9.52AGKK383 pKa = 10.47AILGGKK389 pKa = 9.59GGAKK393 pKa = 9.89LKK395 pKa = 11.21SNDD398 pKa = 3.18FGHH401 pKa = 6.52FFVIAYY407 pKa = 9.41ISAKK411 pKa = 8.32TCYY414 pKa = 9.5PSGFDD419 pKa = 3.61RR420 pKa = 11.84EE421 pKa = 4.42CLKK424 pKa = 10.91SHH426 pKa = 7.18LYY428 pKa = 10.7DD429 pKa = 4.8FYY431 pKa = 11.04TPEE434 pKa = 3.94FDD436 pKa = 5.07GVGLQPISMQEE447 pKa = 3.81LSSEE451 pKa = 4.16NLNFTYY457 pKa = 10.45SEE459 pKa = 4.22PWHH462 pKa = 6.46GNDD465 pKa = 3.07VFGFTEE471 pKa = 4.84RR472 pKa = 11.84YY473 pKa = 8.5NEE475 pKa = 3.72YY476 pKa = 10.6RR477 pKa = 11.84FGKK480 pKa = 10.24DD481 pKa = 2.76RR482 pKa = 11.84ITGDD486 pKa = 3.49MLRR489 pKa = 11.84DD490 pKa = 3.31MDD492 pKa = 4.68FKK494 pKa = 10.54PWYY497 pKa = 9.94LSRR500 pKa = 11.84EE501 pKa = 4.16DD502 pKa = 3.61LTIRR506 pKa = 11.84DD507 pKa = 3.69QDD509 pKa = 3.49VTAQSDD515 pKa = 3.66PLLYY519 pKa = 10.79YY520 pKa = 9.69PINNEE525 pKa = 3.41SSYY528 pKa = 11.07TRR530 pKa = 11.84IFANTSSTLYY540 pKa = 10.28DD541 pKa = 3.52HH542 pKa = 7.53RR543 pKa = 11.84DD544 pKa = 3.32HH545 pKa = 7.27FFMSCYY551 pKa = 10.09FNVSGTRR558 pKa = 11.84PILNLSDD565 pKa = 4.02SLSLGHH571 pKa = 7.41GDD573 pKa = 3.14INLNKK578 pKa = 10.04EE579 pKa = 4.17GLVLSS584 pKa = 4.53
MM1 pKa = 7.61SNNNSQTINISKK13 pKa = 9.6ASKK16 pKa = 9.78YY17 pKa = 10.84SRR19 pKa = 11.84ISLDD23 pKa = 3.46EE24 pKa = 3.98AHH26 pKa = 6.81LTTLDD31 pKa = 3.88FGQIMPIYY39 pKa = 9.79SRR41 pKa = 11.84AIIPGDD47 pKa = 3.91DD48 pKa = 3.68LSIDD52 pKa = 3.7TNVFTRR58 pKa = 11.84TSPLVYY64 pKa = 9.73PSFVNAHH71 pKa = 5.7LTLVSVFVPYY81 pKa = 10.42YY82 pKa = 9.17QLSPFMEE89 pKa = 4.65PFFAGVPNIGSKK101 pKa = 8.27VVKK104 pKa = 10.52LPLLTNHH111 pKa = 7.33IIDD114 pKa = 4.81HH115 pKa = 6.85LFTDD119 pKa = 5.16LAQTLNIGKK128 pKa = 9.59IGQIGVNPDD137 pKa = 4.11IIWTTTDD144 pKa = 2.81NLKK147 pKa = 11.01YY148 pKa = 9.68GTQLNYY154 pKa = 10.4KK155 pKa = 8.78GRR157 pKa = 11.84LMLKK161 pKa = 9.54VLNSLGYY168 pKa = 9.38QVSKK172 pKa = 10.6NVSWKK177 pKa = 10.36SSVDD181 pKa = 3.41NEE183 pKa = 4.5YY184 pKa = 11.14KK185 pKa = 10.69NSPVTLNPLPFISYY199 pKa = 10.0IKK201 pKa = 10.43VYY203 pKa = 10.75KK204 pKa = 10.48DD205 pKa = 2.82WFSSNKK211 pKa = 9.88SFNTSYY217 pKa = 10.82VASILNRR224 pKa = 11.84IQTEE228 pKa = 3.99SDD230 pKa = 3.78FEE232 pKa = 4.1FTPQIINSLFDD243 pKa = 3.98YY244 pKa = 10.91LRR246 pKa = 11.84LLYY249 pKa = 9.73DD250 pKa = 3.18TDD252 pKa = 4.06YY253 pKa = 9.57FTTAWQYY260 pKa = 11.58NNYY263 pKa = 9.56PSDD266 pKa = 3.7SSKK269 pKa = 10.17TSTDD273 pKa = 3.06ILPDD277 pKa = 4.05PSWSGLYY284 pKa = 10.29DD285 pKa = 3.45GNRR288 pKa = 11.84FLINPNEE295 pKa = 3.98NLADD299 pKa = 3.61QSDD302 pKa = 4.27PEE304 pKa = 4.31FLGATTVRR312 pKa = 11.84LLSVLDD318 pKa = 3.54KK319 pKa = 10.72YY320 pKa = 10.89IRR322 pKa = 11.84RR323 pKa = 11.84NSYY326 pKa = 10.99AGTKK330 pKa = 9.28AAARR334 pKa = 11.84VLSRR338 pKa = 11.84FGIKK342 pKa = 8.54STEE345 pKa = 4.11FKK347 pKa = 10.69SQYY350 pKa = 10.89SDD352 pKa = 2.79ILNVDD357 pKa = 4.6SIRR360 pKa = 11.84LNIGDD365 pKa = 3.8VTSTAEE371 pKa = 4.11TTEE374 pKa = 4.2MPLGSYY380 pKa = 9.52AGKK383 pKa = 10.47AILGGKK389 pKa = 9.59GGAKK393 pKa = 9.89LKK395 pKa = 11.21SNDD398 pKa = 3.18FGHH401 pKa = 6.52FFVIAYY407 pKa = 9.41ISAKK411 pKa = 8.32TCYY414 pKa = 9.5PSGFDD419 pKa = 3.61RR420 pKa = 11.84EE421 pKa = 4.42CLKK424 pKa = 10.91SHH426 pKa = 7.18LYY428 pKa = 10.7DD429 pKa = 4.8FYY431 pKa = 11.04TPEE434 pKa = 3.94FDD436 pKa = 5.07GVGLQPISMQEE447 pKa = 3.81LSSEE451 pKa = 4.16NLNFTYY457 pKa = 10.45SEE459 pKa = 4.22PWHH462 pKa = 6.46GNDD465 pKa = 3.07VFGFTEE471 pKa = 4.84RR472 pKa = 11.84YY473 pKa = 8.5NEE475 pKa = 3.72YY476 pKa = 10.6RR477 pKa = 11.84FGKK480 pKa = 10.24DD481 pKa = 2.76RR482 pKa = 11.84ITGDD486 pKa = 3.49MLRR489 pKa = 11.84DD490 pKa = 3.31MDD492 pKa = 4.68FKK494 pKa = 10.54PWYY497 pKa = 9.94LSRR500 pKa = 11.84EE501 pKa = 4.16DD502 pKa = 3.61LTIRR506 pKa = 11.84DD507 pKa = 3.69QDD509 pKa = 3.49VTAQSDD515 pKa = 3.66PLLYY519 pKa = 10.79YY520 pKa = 9.69PINNEE525 pKa = 3.41SSYY528 pKa = 11.07TRR530 pKa = 11.84IFANTSSTLYY540 pKa = 10.28DD541 pKa = 3.52HH542 pKa = 7.53RR543 pKa = 11.84DD544 pKa = 3.32HH545 pKa = 7.27FFMSCYY551 pKa = 10.09FNVSGTRR558 pKa = 11.84PILNLSDD565 pKa = 4.02SLSLGHH571 pKa = 7.41GDD573 pKa = 3.14INLNKK578 pKa = 10.04EE579 pKa = 4.17GLVLSS584 pKa = 4.53
Molecular weight: 66.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W3U8|A0A4P8W3U8_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_45 OX=2584778 PE=3 SV=1
MM1 pKa = 7.72SYY3 pKa = 9.2TFKK6 pKa = 11.16VEE8 pKa = 4.05NKK10 pKa = 6.67THH12 pKa = 6.01YY13 pKa = 10.48FGEE16 pKa = 4.16NTKK19 pKa = 10.64LYY21 pKa = 10.74NNIVNPRR28 pKa = 11.84SYY30 pKa = 11.26AALHH34 pKa = 5.96SKK36 pKa = 9.48QLSYY40 pKa = 10.79YY41 pKa = 8.23VRR43 pKa = 11.84SYY45 pKa = 12.0YY46 pKa = 10.67EE47 pKa = 3.81FLKK50 pKa = 10.23TKK52 pKa = 9.72YY53 pKa = 10.69LNGKK57 pKa = 6.9TMFLTFTYY65 pKa = 10.02CDD67 pKa = 3.18NALPILYY74 pKa = 9.51GIPTVDD80 pKa = 3.47NNNIRR85 pKa = 11.84YY86 pKa = 9.01LLRR89 pKa = 11.84NSYY92 pKa = 10.14LPKK95 pKa = 9.92FVASKK100 pKa = 9.81GYY102 pKa = 9.97KK103 pKa = 7.17MSYY106 pKa = 9.23MIVPEE111 pKa = 4.08FGEE114 pKa = 4.79GKK116 pKa = 10.21GSRR119 pKa = 11.84GVGNNPHH126 pKa = 5.96FHH128 pKa = 6.89CLLHH132 pKa = 6.47FYY134 pKa = 10.67KK135 pKa = 10.18EE136 pKa = 4.21TGEE139 pKa = 4.07PFAYY143 pKa = 10.31ADD145 pKa = 4.59GVDD148 pKa = 3.86IYY150 pKa = 11.9NKK152 pKa = 9.89ILNLWQGTNPDD163 pKa = 3.32GTITRR168 pKa = 11.84DD169 pKa = 3.11PLKK172 pKa = 10.21YY173 pKa = 10.23HH174 pKa = 6.82FGMVQASANGLYY186 pKa = 9.86IDD188 pKa = 5.27DD189 pKa = 4.75YY190 pKa = 10.71KK191 pKa = 11.34ACSYY195 pKa = 9.92VSKK198 pKa = 9.88YY199 pKa = 7.85TSKK202 pKa = 11.11DD203 pKa = 2.7INTRR207 pKa = 11.84TKK209 pKa = 9.95ISQIEE214 pKa = 3.99KK215 pKa = 10.4AIEE218 pKa = 4.01LDD220 pKa = 3.62CFHH223 pKa = 7.02YY224 pKa = 9.95CTDD227 pKa = 4.26SSICDD232 pKa = 3.08IMNSLNVSDD241 pKa = 4.64SLGSYY246 pKa = 7.74LHH248 pKa = 6.73KK249 pKa = 10.59KK250 pKa = 9.27YY251 pKa = 10.91LSDD254 pKa = 4.07DD255 pKa = 3.42SPYY258 pKa = 10.97RR259 pKa = 11.84KK260 pKa = 10.24DD261 pKa = 3.02FEE263 pKa = 4.53KK264 pKa = 10.53RR265 pKa = 11.84VKK267 pKa = 10.54QMSAVIFADD276 pKa = 3.0IKK278 pKa = 10.96NRR280 pKa = 11.84YY281 pKa = 7.47LPKK284 pKa = 9.77VFMSKK289 pKa = 10.46GYY291 pKa = 10.98GDD293 pKa = 3.66YY294 pKa = 11.37ALNFVNEE301 pKa = 4.36DD302 pKa = 3.28FKK304 pKa = 11.83LPIPQSNPNAKK315 pKa = 9.44VQGTIFIDD323 pKa = 3.75LPLYY327 pKa = 10.53LYY329 pKa = 10.49RR330 pKa = 11.84KK331 pKa = 9.93LFTDD335 pKa = 2.89VHH337 pKa = 7.76KK338 pKa = 10.81DD339 pKa = 3.09EE340 pKa = 5.26KK341 pKa = 11.47GNVIYY346 pKa = 10.74VLNDD350 pKa = 3.18TGKK353 pKa = 9.42HH354 pKa = 4.25YY355 pKa = 11.34KK356 pKa = 7.94NTRR359 pKa = 11.84LLKK362 pKa = 10.77DD363 pKa = 3.0IDD365 pKa = 3.85KK366 pKa = 9.62TCNDD370 pKa = 3.8VACLISDD377 pKa = 4.11NFDD380 pKa = 3.36MPLDD384 pKa = 3.77QLSSLSYY391 pKa = 10.54DD392 pKa = 3.32YY393 pKa = 11.51SLYY396 pKa = 9.61KK397 pKa = 10.2HH398 pKa = 6.66IYY400 pKa = 7.11EE401 pKa = 4.45HH402 pKa = 6.24HH403 pKa = 6.62TFDD406 pKa = 3.84IFQPHH411 pKa = 6.34EE412 pKa = 4.49LDD414 pKa = 3.4PLGDD418 pKa = 3.53YY419 pKa = 10.47TRR421 pKa = 11.84FLVSGFEE428 pKa = 4.18TEE430 pKa = 4.26SFEE433 pKa = 4.4EE434 pKa = 4.96FYY436 pKa = 10.77FVKK439 pKa = 10.17KK440 pKa = 10.47RR441 pKa = 11.84KK442 pKa = 9.17LLQSRR447 pKa = 11.84LTYY450 pKa = 10.1EE451 pKa = 3.75FHH453 pKa = 7.63PYY455 pKa = 9.17FAPLIPYY462 pKa = 9.08FRR464 pKa = 11.84IIDD467 pKa = 3.8NFVDD471 pKa = 3.67NLNSEE476 pKa = 4.23NDD478 pKa = 3.12KK479 pKa = 11.27ALYY482 pKa = 10.42DD483 pKa = 3.72NYY485 pKa = 11.29LEE487 pKa = 4.61IIRR490 pKa = 11.84LRR492 pKa = 11.84KK493 pKa = 9.11VHH495 pKa = 6.43NKK497 pKa = 8.19MLL499 pKa = 4.26
MM1 pKa = 7.72SYY3 pKa = 9.2TFKK6 pKa = 11.16VEE8 pKa = 4.05NKK10 pKa = 6.67THH12 pKa = 6.01YY13 pKa = 10.48FGEE16 pKa = 4.16NTKK19 pKa = 10.64LYY21 pKa = 10.74NNIVNPRR28 pKa = 11.84SYY30 pKa = 11.26AALHH34 pKa = 5.96SKK36 pKa = 9.48QLSYY40 pKa = 10.79YY41 pKa = 8.23VRR43 pKa = 11.84SYY45 pKa = 12.0YY46 pKa = 10.67EE47 pKa = 3.81FLKK50 pKa = 10.23TKK52 pKa = 9.72YY53 pKa = 10.69LNGKK57 pKa = 6.9TMFLTFTYY65 pKa = 10.02CDD67 pKa = 3.18NALPILYY74 pKa = 9.51GIPTVDD80 pKa = 3.47NNNIRR85 pKa = 11.84YY86 pKa = 9.01LLRR89 pKa = 11.84NSYY92 pKa = 10.14LPKK95 pKa = 9.92FVASKK100 pKa = 9.81GYY102 pKa = 9.97KK103 pKa = 7.17MSYY106 pKa = 9.23MIVPEE111 pKa = 4.08FGEE114 pKa = 4.79GKK116 pKa = 10.21GSRR119 pKa = 11.84GVGNNPHH126 pKa = 5.96FHH128 pKa = 6.89CLLHH132 pKa = 6.47FYY134 pKa = 10.67KK135 pKa = 10.18EE136 pKa = 4.21TGEE139 pKa = 4.07PFAYY143 pKa = 10.31ADD145 pKa = 4.59GVDD148 pKa = 3.86IYY150 pKa = 11.9NKK152 pKa = 9.89ILNLWQGTNPDD163 pKa = 3.32GTITRR168 pKa = 11.84DD169 pKa = 3.11PLKK172 pKa = 10.21YY173 pKa = 10.23HH174 pKa = 6.82FGMVQASANGLYY186 pKa = 9.86IDD188 pKa = 5.27DD189 pKa = 4.75YY190 pKa = 10.71KK191 pKa = 11.34ACSYY195 pKa = 9.92VSKK198 pKa = 9.88YY199 pKa = 7.85TSKK202 pKa = 11.11DD203 pKa = 2.7INTRR207 pKa = 11.84TKK209 pKa = 9.95ISQIEE214 pKa = 3.99KK215 pKa = 10.4AIEE218 pKa = 4.01LDD220 pKa = 3.62CFHH223 pKa = 7.02YY224 pKa = 9.95CTDD227 pKa = 4.26SSICDD232 pKa = 3.08IMNSLNVSDD241 pKa = 4.64SLGSYY246 pKa = 7.74LHH248 pKa = 6.73KK249 pKa = 10.59KK250 pKa = 9.27YY251 pKa = 10.91LSDD254 pKa = 4.07DD255 pKa = 3.42SPYY258 pKa = 10.97RR259 pKa = 11.84KK260 pKa = 10.24DD261 pKa = 3.02FEE263 pKa = 4.53KK264 pKa = 10.53RR265 pKa = 11.84VKK267 pKa = 10.54QMSAVIFADD276 pKa = 3.0IKK278 pKa = 10.96NRR280 pKa = 11.84YY281 pKa = 7.47LPKK284 pKa = 9.77VFMSKK289 pKa = 10.46GYY291 pKa = 10.98GDD293 pKa = 3.66YY294 pKa = 11.37ALNFVNEE301 pKa = 4.36DD302 pKa = 3.28FKK304 pKa = 11.83LPIPQSNPNAKK315 pKa = 9.44VQGTIFIDD323 pKa = 3.75LPLYY327 pKa = 10.53LYY329 pKa = 10.49RR330 pKa = 11.84KK331 pKa = 9.93LFTDD335 pKa = 2.89VHH337 pKa = 7.76KK338 pKa = 10.81DD339 pKa = 3.09EE340 pKa = 5.26KK341 pKa = 11.47GNVIYY346 pKa = 10.74VLNDD350 pKa = 3.18TGKK353 pKa = 9.42HH354 pKa = 4.25YY355 pKa = 11.34KK356 pKa = 7.94NTRR359 pKa = 11.84LLKK362 pKa = 10.77DD363 pKa = 3.0IDD365 pKa = 3.85KK366 pKa = 9.62TCNDD370 pKa = 3.8VACLISDD377 pKa = 4.11NFDD380 pKa = 3.36MPLDD384 pKa = 3.77QLSSLSYY391 pKa = 10.54DD392 pKa = 3.32YY393 pKa = 11.51SLYY396 pKa = 9.61KK397 pKa = 10.2HH398 pKa = 6.66IYY400 pKa = 7.11EE401 pKa = 4.45HH402 pKa = 6.24HH403 pKa = 6.62TFDD406 pKa = 3.84IFQPHH411 pKa = 6.34EE412 pKa = 4.49LDD414 pKa = 3.4PLGDD418 pKa = 3.53YY419 pKa = 10.47TRR421 pKa = 11.84FLVSGFEE428 pKa = 4.18TEE430 pKa = 4.26SFEE433 pKa = 4.4EE434 pKa = 4.96FYY436 pKa = 10.77FVKK439 pKa = 10.17KK440 pKa = 10.47RR441 pKa = 11.84KK442 pKa = 9.17LLQSRR447 pKa = 11.84LTYY450 pKa = 10.1EE451 pKa = 3.75FHH453 pKa = 7.63PYY455 pKa = 9.17FAPLIPYY462 pKa = 9.08FRR464 pKa = 11.84IIDD467 pKa = 3.8NFVDD471 pKa = 3.67NLNSEE476 pKa = 4.23NDD478 pKa = 3.12KK479 pKa = 11.27ALYY482 pKa = 10.42DD483 pKa = 3.72NYY485 pKa = 11.29LEE487 pKa = 4.61IIRR490 pKa = 11.84LRR492 pKa = 11.84KK493 pKa = 9.11VHH495 pKa = 6.43NKK497 pKa = 8.19MLL499 pKa = 4.26
Molecular weight: 58.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1696 |
101 |
584 |
339.2 |
38.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.894 ± 1.149 | 1.179 ± 0.539 |
6.958 ± 0.319 | 4.127 ± 0.576 |
5.483 ± 0.645 | 5.425 ± 0.461 |
2.358 ± 0.376 | 6.309 ± 0.39 |
6.663 ± 0.944 | 10.436 ± 0.351 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.44 | 7.724 ± 0.478 |
4.068 ± 0.837 | 3.243 ± 0.927 |
3.656 ± 0.134 | 9.198 ± 1.266 |
5.307 ± 0.864 | 4.422 ± 0.624 |
0.884 ± 0.286 | 5.66 ± 1.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
