
Pontibacillus halophilus JSM 076056 = DSM 19796
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Pontibacillus; Pontibacillus halophilus
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
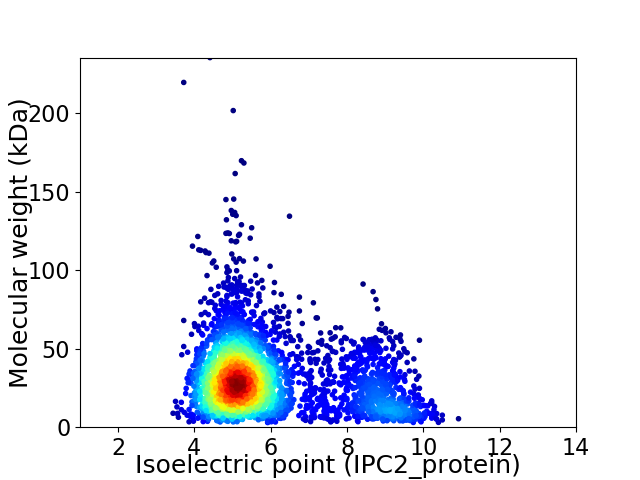
Virtual 2D-PAGE plot for 3559 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
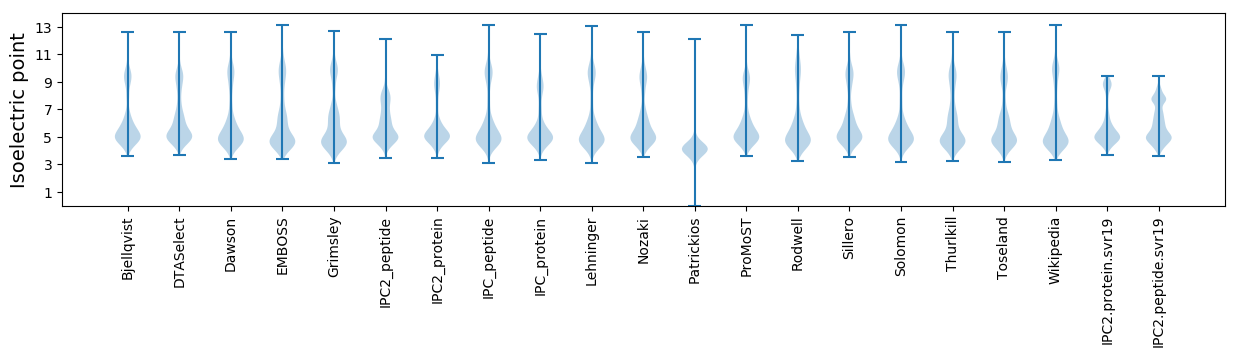
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A5GEA1|A0A0A5GEA1_9BACI NADH-dependent butanol dehydrogenase A OS=Pontibacillus halophilus JSM 076056 = DSM 19796 OX=1385510 GN=N781_07500 PE=4 SV=1
MM1 pKa = 7.45AFVNRR6 pKa = 11.84FSTLDD11 pKa = 3.4CGMMTFTGNTLGLSQEE27 pKa = 4.33LNLNEE32 pKa = 4.24AGTRR36 pKa = 11.84GSIGAFITLDD46 pKa = 3.36NTSQVPTFPPGTTLNYY62 pKa = 10.16QEE64 pKa = 4.71NSSSAEE70 pKa = 3.81LGFAPGSNTVLYY82 pKa = 10.91AEE84 pKa = 5.47LIWGGNYY91 pKa = 7.55LTRR94 pKa = 11.84DD95 pKa = 3.25EE96 pKa = 6.13DD97 pKa = 3.56ITSEE101 pKa = 3.87IDD103 pKa = 3.18NDD105 pKa = 4.11VLFTDD110 pKa = 4.64PLGVAHH116 pKa = 7.52PISPDD121 pKa = 3.39PATSNQFTFTIGGVTRR137 pKa = 11.84GFYY140 pKa = 9.49MRR142 pKa = 11.84SNDD145 pKa = 3.14VTSIVQSAAEE155 pKa = 4.08GTYY158 pKa = 9.5TCGSVPGLLDD168 pKa = 3.66PLIASTGDD176 pKa = 3.27TNHH179 pKa = 7.11AGWTLAVVYY188 pKa = 9.1GNPEE192 pKa = 3.81LPNRR196 pKa = 11.84SMNLFVGGEE205 pKa = 4.39GIVFSQGTPIIDD217 pKa = 3.34IAVSGFMTPPSGDD230 pKa = 2.59VDD232 pKa = 3.5ARR234 pKa = 11.84LLISAQEE241 pKa = 3.93GDD243 pKa = 3.68ANIPGDD249 pKa = 3.6QALFGPDD256 pKa = 3.78SLTLTNLSGPNNPTFNFSASQINGDD281 pKa = 4.41DD282 pKa = 3.93GLVDD286 pKa = 3.37TSGTFGTRR294 pKa = 11.84NQDD297 pKa = 2.74AATQTNIVAGRR308 pKa = 11.84QGWDD312 pKa = 2.85ITNVSGLNYY321 pKa = 10.25LPNNQTTAVFRR332 pKa = 11.84FTSTGDD338 pKa = 3.04AYY340 pKa = 10.65MPNALGIQIDD350 pKa = 4.21EE351 pKa = 4.48GDD353 pKa = 4.07ADD355 pKa = 3.99PVLVKK360 pKa = 10.04TVDD363 pKa = 3.15KK364 pKa = 10.7PFAVVNDD371 pKa = 3.41VLTYY375 pKa = 10.36IITITNEE382 pKa = 3.79GVIEE386 pKa = 4.22ANNVVFIDD394 pKa = 4.79EE395 pKa = 4.81IPPGTLFVEE404 pKa = 4.7DD405 pKa = 3.83SLYY408 pKa = 10.26IDD410 pKa = 4.92GVQFPGVNPQFGVPLPNIDD429 pKa = 3.51VGDD432 pKa = 3.84TVTVFFQVIVRR443 pKa = 11.84RR444 pKa = 11.84CDD446 pKa = 3.39CFVQNRR452 pKa = 11.84AEE454 pKa = 4.43LEE456 pKa = 4.19FSCGKK461 pKa = 9.82VAEE464 pKa = 4.7SNLVTSTVCFQCYY477 pKa = 9.37CGSGFICC484 pKa = 5.34
MM1 pKa = 7.45AFVNRR6 pKa = 11.84FSTLDD11 pKa = 3.4CGMMTFTGNTLGLSQEE27 pKa = 4.33LNLNEE32 pKa = 4.24AGTRR36 pKa = 11.84GSIGAFITLDD46 pKa = 3.36NTSQVPTFPPGTTLNYY62 pKa = 10.16QEE64 pKa = 4.71NSSSAEE70 pKa = 3.81LGFAPGSNTVLYY82 pKa = 10.91AEE84 pKa = 5.47LIWGGNYY91 pKa = 7.55LTRR94 pKa = 11.84DD95 pKa = 3.25EE96 pKa = 6.13DD97 pKa = 3.56ITSEE101 pKa = 3.87IDD103 pKa = 3.18NDD105 pKa = 4.11VLFTDD110 pKa = 4.64PLGVAHH116 pKa = 7.52PISPDD121 pKa = 3.39PATSNQFTFTIGGVTRR137 pKa = 11.84GFYY140 pKa = 9.49MRR142 pKa = 11.84SNDD145 pKa = 3.14VTSIVQSAAEE155 pKa = 4.08GTYY158 pKa = 9.5TCGSVPGLLDD168 pKa = 3.66PLIASTGDD176 pKa = 3.27TNHH179 pKa = 7.11AGWTLAVVYY188 pKa = 9.1GNPEE192 pKa = 3.81LPNRR196 pKa = 11.84SMNLFVGGEE205 pKa = 4.39GIVFSQGTPIIDD217 pKa = 3.34IAVSGFMTPPSGDD230 pKa = 2.59VDD232 pKa = 3.5ARR234 pKa = 11.84LLISAQEE241 pKa = 3.93GDD243 pKa = 3.68ANIPGDD249 pKa = 3.6QALFGPDD256 pKa = 3.78SLTLTNLSGPNNPTFNFSASQINGDD281 pKa = 4.41DD282 pKa = 3.93GLVDD286 pKa = 3.37TSGTFGTRR294 pKa = 11.84NQDD297 pKa = 2.74AATQTNIVAGRR308 pKa = 11.84QGWDD312 pKa = 2.85ITNVSGLNYY321 pKa = 10.25LPNNQTTAVFRR332 pKa = 11.84FTSTGDD338 pKa = 3.04AYY340 pKa = 10.65MPNALGIQIDD350 pKa = 4.21EE351 pKa = 4.48GDD353 pKa = 4.07ADD355 pKa = 3.99PVLVKK360 pKa = 10.04TVDD363 pKa = 3.15KK364 pKa = 10.7PFAVVNDD371 pKa = 3.41VLTYY375 pKa = 10.36IITITNEE382 pKa = 3.79GVIEE386 pKa = 4.22ANNVVFIDD394 pKa = 4.79EE395 pKa = 4.81IPPGTLFVEE404 pKa = 4.7DD405 pKa = 3.83SLYY408 pKa = 10.26IDD410 pKa = 4.92GVQFPGVNPQFGVPLPNIDD429 pKa = 3.51VGDD432 pKa = 3.84TVTVFFQVIVRR443 pKa = 11.84RR444 pKa = 11.84CDD446 pKa = 3.39CFVQNRR452 pKa = 11.84AEE454 pKa = 4.43LEE456 pKa = 4.19FSCGKK461 pKa = 9.82VAEE464 pKa = 4.7SNLVTSTVCFQCYY477 pKa = 9.37CGSGFICC484 pKa = 5.34
Molecular weight: 51.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A5GP05|A0A0A5GP05_9BACI Aspartate racemase OS=Pontibacillus halophilus JSM 076056 = DSM 19796 OX=1385510 GN=N781_00460 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.1NGRR28 pKa = 11.84EE29 pKa = 3.63VLKK32 pKa = 10.46RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.96GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 10.1NGRR28 pKa = 11.84EE29 pKa = 3.63VLKK32 pKa = 10.46RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.96GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015912 |
26 |
2105 |
285.4 |
32.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.855 ± 0.038 | 0.592 ± 0.013 |
5.276 ± 0.037 | 8.051 ± 0.059 |
4.371 ± 0.038 | 7.041 ± 0.041 |
2.343 ± 0.025 | 6.743 ± 0.037 |
5.672 ± 0.033 | 9.755 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.857 ± 0.019 | 3.981 ± 0.028 |
3.659 ± 0.024 | 4.188 ± 0.027 |
4.398 ± 0.032 | 6.192 ± 0.032 |
5.727 ± 0.03 | 7.516 ± 0.036 |
1.085 ± 0.018 | 3.696 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
