
Duganella sp. CF402
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Duganella; unclassified Duganella
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
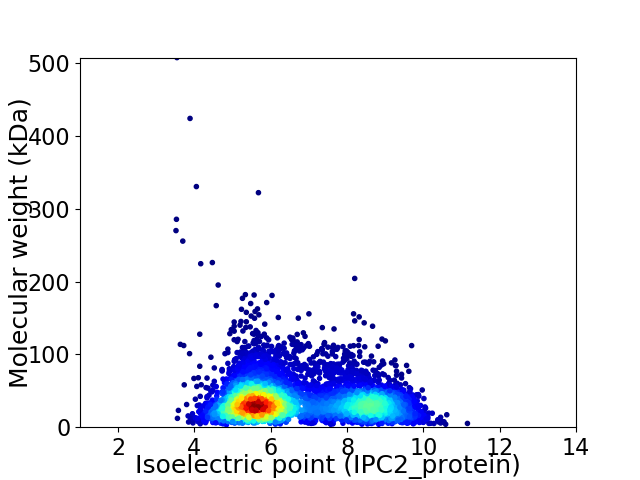
Virtual 2D-PAGE plot for 5435 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8AE70|A0A1H8AE70_9BURK Cytochrome c554 and c-prime OS=Duganella sp. CF402 OX=1855289 GN=SAMN05216319_4480 PE=4 SV=1
MM1 pKa = 7.64TSPALLSVSSSDD13 pKa = 2.95NSTIVLHH20 pKa = 6.52FDD22 pKa = 3.4RR23 pKa = 11.84NVFAGDD29 pKa = 3.43GSIVVSDD36 pKa = 4.84GYY38 pKa = 11.52GQTFLSDD45 pKa = 2.69GGLRR49 pKa = 11.84TRR51 pKa = 11.84IVNASDD57 pKa = 3.37QRR59 pKa = 11.84ALDD62 pKa = 4.09ASDD65 pKa = 3.85SALTYY70 pKa = 10.22SGQNITIHH78 pKa = 7.01LSTPLKK84 pKa = 10.79NGLNYY89 pKa = 10.25SITMSGGTVIDD100 pKa = 5.0DD101 pKa = 4.03SDD103 pKa = 4.63DD104 pKa = 4.29GIARR108 pKa = 11.84ISSPNLFKK116 pKa = 10.42FTASGSAATVPTPSAVIGSTLHH138 pKa = 5.53FTDD141 pKa = 4.13TGTSSSDD148 pKa = 3.65YY149 pKa = 8.87ITASASQVMTGTYY162 pKa = 9.5TGTLGASDD170 pKa = 4.49FVQVSLDD177 pKa = 4.12NGASWHH183 pKa = 6.24KK184 pKa = 10.18ATVNASAKK192 pKa = 7.34TWSYY196 pKa = 10.4TGEE199 pKa = 3.98IDD201 pKa = 5.73LDD203 pKa = 3.94NLTDD207 pKa = 3.93GAGDD211 pKa = 3.95ALDD214 pKa = 3.95GTLLARR220 pKa = 11.84IGNTSGGSSGTASHH234 pKa = 6.47TYY236 pKa = 9.95VYY238 pKa = 10.52NSHH241 pKa = 6.52AVEE244 pKa = 4.38IDD246 pKa = 3.43VSSSFSFSADD256 pKa = 2.85TGTSSTDD263 pKa = 4.02LITKK267 pKa = 8.01TAAQTISGSYY277 pKa = 9.85SGSLLAGYY285 pKa = 6.8TLQVSVDD292 pKa = 3.87GGANWINASASAGSWHH308 pKa = 5.58TTGTVNLKK316 pKa = 10.43SGANEE321 pKa = 3.59VQARR325 pKa = 11.84VIDD328 pKa = 3.93TAGNTSGVASADD340 pKa = 3.49YY341 pKa = 11.02KK342 pKa = 11.21LITSTVSLAGHH353 pKa = 7.46ALTLAEE359 pKa = 4.98GSDD362 pKa = 3.99TGPSATDD369 pKa = 4.88GITRR373 pKa = 11.84SASKK377 pKa = 9.23VTLNVAGLHH386 pKa = 6.02GFHH389 pKa = 7.12AGDD392 pKa = 4.44TIQIVDD398 pKa = 3.99TSHH401 pKa = 6.52SSAVVGSYY409 pKa = 10.84VILDD413 pKa = 3.44ADD415 pKa = 4.59LYY417 pKa = 11.46YY418 pKa = 11.17GDD420 pKa = 5.79DD421 pKa = 3.73YY422 pKa = 11.38FTISKK427 pKa = 9.24FNPTARR433 pKa = 11.84TTVDD437 pKa = 2.86INLDD441 pKa = 3.52TMADD445 pKa = 3.83GAHH448 pKa = 5.99TLAARR453 pKa = 11.84IVDD456 pKa = 3.8VAGNTAAASGTTAVTVDD473 pKa = 3.36GKK475 pKa = 11.12VPLMLSSAPLEE486 pKa = 4.56DD487 pKa = 3.3ATGVSTGLTKK497 pKa = 10.16LTFTFNEE504 pKa = 4.23NIAIEE509 pKa = 4.86DD510 pKa = 3.84GTIVTITDD518 pKa = 4.04DD519 pKa = 3.83NNSANSQEE527 pKa = 4.04ITLSSTAASGNKK539 pKa = 8.23LTINLSSALTSGTSYY554 pKa = 10.37TVIGAIVTDD563 pKa = 4.09LAGNDD568 pKa = 3.86GVTGDD573 pKa = 3.85TPLLHH578 pKa = 6.42FTTEE582 pKa = 3.56GTYY585 pKa = 10.79AGGSTPDD592 pKa = 3.51SVAADD597 pKa = 3.91YY598 pKa = 11.71VDD600 pKa = 4.44TAPAWDD606 pKa = 3.98TDD608 pKa = 4.05SGSDD612 pKa = 3.3AHH614 pKa = 6.97NDD616 pKa = 3.09GVTSNTRR623 pKa = 11.84ISVTGAPLTEE633 pKa = 3.51WHH635 pKa = 6.54YY636 pKa = 11.45SVNGTTYY643 pKa = 10.59LGSGDD648 pKa = 4.4GFDD651 pKa = 5.68LEE653 pKa = 5.36DD654 pKa = 3.39GTYY657 pKa = 9.31TAGQIQVWQTVDD669 pKa = 3.52GVDD672 pKa = 3.81SAHH675 pKa = 5.94TSIGKK680 pKa = 7.87TLVIDD685 pKa = 4.04TLAPTAVAHH694 pKa = 6.16GVPEE698 pKa = 4.37SFSSGDD704 pKa = 3.43SSIEE708 pKa = 3.8GLVMGTTDD716 pKa = 3.04VAEE719 pKa = 4.39EE720 pKa = 4.03FVEE723 pKa = 4.52VTFDD727 pKa = 4.7HH728 pKa = 6.73GVTWVQAEE736 pKa = 4.34TTYY739 pKa = 11.49VNADD743 pKa = 3.26YY744 pKa = 9.44STWSLAGISASLGNNYY760 pKa = 9.45GLRR763 pKa = 11.84LSDD766 pKa = 3.21TAGNISTFNALSATNPVYY784 pKa = 10.87YY785 pKa = 10.42LADD788 pKa = 3.88GGVTYY793 pKa = 10.86SHH795 pKa = 7.22ASDD798 pKa = 5.51DD799 pKa = 3.91YY800 pKa = 8.31TTVFGGSGADD810 pKa = 3.61TITVGAHH817 pKa = 5.71ALVAGGDD824 pKa = 3.63GDD826 pKa = 5.59IITTGANSAIITRR839 pKa = 11.84ASSTVVTGGGTNVVEE854 pKa = 4.67ADD856 pKa = 3.7SNAHH860 pKa = 5.61ITTGSGTDD868 pKa = 3.69SITLTSLTGAVLAAGSGTDD887 pKa = 3.47TLILEE892 pKa = 4.57FDD894 pKa = 3.82GSFTLGTALGSSSGIDD910 pKa = 3.08IIDD913 pKa = 4.07FGSDD917 pKa = 2.98GTGHH921 pKa = 6.01MNILTSASVHH931 pKa = 5.99NLTDD935 pKa = 3.71SDD937 pKa = 3.91QLTVNDD943 pKa = 4.24SGGGATTMTLEE954 pKa = 4.2SLVWAHH960 pKa = 6.54TGTSGAYY967 pKa = 9.04EE968 pKa = 3.96IYY970 pKa = 10.52KK971 pKa = 8.71NTVSGGDD978 pKa = 3.4GTVVVLIANTIDD990 pKa = 3.31VTLDD994 pKa = 2.94TFTAA998 pKa = 3.89
MM1 pKa = 7.64TSPALLSVSSSDD13 pKa = 2.95NSTIVLHH20 pKa = 6.52FDD22 pKa = 3.4RR23 pKa = 11.84NVFAGDD29 pKa = 3.43GSIVVSDD36 pKa = 4.84GYY38 pKa = 11.52GQTFLSDD45 pKa = 2.69GGLRR49 pKa = 11.84TRR51 pKa = 11.84IVNASDD57 pKa = 3.37QRR59 pKa = 11.84ALDD62 pKa = 4.09ASDD65 pKa = 3.85SALTYY70 pKa = 10.22SGQNITIHH78 pKa = 7.01LSTPLKK84 pKa = 10.79NGLNYY89 pKa = 10.25SITMSGGTVIDD100 pKa = 5.0DD101 pKa = 4.03SDD103 pKa = 4.63DD104 pKa = 4.29GIARR108 pKa = 11.84ISSPNLFKK116 pKa = 10.42FTASGSAATVPTPSAVIGSTLHH138 pKa = 5.53FTDD141 pKa = 4.13TGTSSSDD148 pKa = 3.65YY149 pKa = 8.87ITASASQVMTGTYY162 pKa = 9.5TGTLGASDD170 pKa = 4.49FVQVSLDD177 pKa = 4.12NGASWHH183 pKa = 6.24KK184 pKa = 10.18ATVNASAKK192 pKa = 7.34TWSYY196 pKa = 10.4TGEE199 pKa = 3.98IDD201 pKa = 5.73LDD203 pKa = 3.94NLTDD207 pKa = 3.93GAGDD211 pKa = 3.95ALDD214 pKa = 3.95GTLLARR220 pKa = 11.84IGNTSGGSSGTASHH234 pKa = 6.47TYY236 pKa = 9.95VYY238 pKa = 10.52NSHH241 pKa = 6.52AVEE244 pKa = 4.38IDD246 pKa = 3.43VSSSFSFSADD256 pKa = 2.85TGTSSTDD263 pKa = 4.02LITKK267 pKa = 8.01TAAQTISGSYY277 pKa = 9.85SGSLLAGYY285 pKa = 6.8TLQVSVDD292 pKa = 3.87GGANWINASASAGSWHH308 pKa = 5.58TTGTVNLKK316 pKa = 10.43SGANEE321 pKa = 3.59VQARR325 pKa = 11.84VIDD328 pKa = 3.93TAGNTSGVASADD340 pKa = 3.49YY341 pKa = 11.02KK342 pKa = 11.21LITSTVSLAGHH353 pKa = 7.46ALTLAEE359 pKa = 4.98GSDD362 pKa = 3.99TGPSATDD369 pKa = 4.88GITRR373 pKa = 11.84SASKK377 pKa = 9.23VTLNVAGLHH386 pKa = 6.02GFHH389 pKa = 7.12AGDD392 pKa = 4.44TIQIVDD398 pKa = 3.99TSHH401 pKa = 6.52SSAVVGSYY409 pKa = 10.84VILDD413 pKa = 3.44ADD415 pKa = 4.59LYY417 pKa = 11.46YY418 pKa = 11.17GDD420 pKa = 5.79DD421 pKa = 3.73YY422 pKa = 11.38FTISKK427 pKa = 9.24FNPTARR433 pKa = 11.84TTVDD437 pKa = 2.86INLDD441 pKa = 3.52TMADD445 pKa = 3.83GAHH448 pKa = 5.99TLAARR453 pKa = 11.84IVDD456 pKa = 3.8VAGNTAAASGTTAVTVDD473 pKa = 3.36GKK475 pKa = 11.12VPLMLSSAPLEE486 pKa = 4.56DD487 pKa = 3.3ATGVSTGLTKK497 pKa = 10.16LTFTFNEE504 pKa = 4.23NIAIEE509 pKa = 4.86DD510 pKa = 3.84GTIVTITDD518 pKa = 4.04DD519 pKa = 3.83NNSANSQEE527 pKa = 4.04ITLSSTAASGNKK539 pKa = 8.23LTINLSSALTSGTSYY554 pKa = 10.37TVIGAIVTDD563 pKa = 4.09LAGNDD568 pKa = 3.86GVTGDD573 pKa = 3.85TPLLHH578 pKa = 6.42FTTEE582 pKa = 3.56GTYY585 pKa = 10.79AGGSTPDD592 pKa = 3.51SVAADD597 pKa = 3.91YY598 pKa = 11.71VDD600 pKa = 4.44TAPAWDD606 pKa = 3.98TDD608 pKa = 4.05SGSDD612 pKa = 3.3AHH614 pKa = 6.97NDD616 pKa = 3.09GVTSNTRR623 pKa = 11.84ISVTGAPLTEE633 pKa = 3.51WHH635 pKa = 6.54YY636 pKa = 11.45SVNGTTYY643 pKa = 10.59LGSGDD648 pKa = 4.4GFDD651 pKa = 5.68LEE653 pKa = 5.36DD654 pKa = 3.39GTYY657 pKa = 9.31TAGQIQVWQTVDD669 pKa = 3.52GVDD672 pKa = 3.81SAHH675 pKa = 5.94TSIGKK680 pKa = 7.87TLVIDD685 pKa = 4.04TLAPTAVAHH694 pKa = 6.16GVPEE698 pKa = 4.37SFSSGDD704 pKa = 3.43SSIEE708 pKa = 3.8GLVMGTTDD716 pKa = 3.04VAEE719 pKa = 4.39EE720 pKa = 4.03FVEE723 pKa = 4.52VTFDD727 pKa = 4.7HH728 pKa = 6.73GVTWVQAEE736 pKa = 4.34TTYY739 pKa = 11.49VNADD743 pKa = 3.26YY744 pKa = 9.44STWSLAGISASLGNNYY760 pKa = 9.45GLRR763 pKa = 11.84LSDD766 pKa = 3.21TAGNISTFNALSATNPVYY784 pKa = 10.87YY785 pKa = 10.42LADD788 pKa = 3.88GGVTYY793 pKa = 10.86SHH795 pKa = 7.22ASDD798 pKa = 5.51DD799 pKa = 3.91YY800 pKa = 8.31TTVFGGSGADD810 pKa = 3.61TITVGAHH817 pKa = 5.71ALVAGGDD824 pKa = 3.63GDD826 pKa = 5.59IITTGANSAIITRR839 pKa = 11.84ASSTVVTGGGTNVVEE854 pKa = 4.67ADD856 pKa = 3.7SNAHH860 pKa = 5.61ITTGSGTDD868 pKa = 3.69SITLTSLTGAVLAAGSGTDD887 pKa = 3.47TLILEE892 pKa = 4.57FDD894 pKa = 3.82GSFTLGTALGSSSGIDD910 pKa = 3.08IIDD913 pKa = 4.07FGSDD917 pKa = 2.98GTGHH921 pKa = 6.01MNILTSASVHH931 pKa = 5.99NLTDD935 pKa = 3.71SDD937 pKa = 3.91QLTVNDD943 pKa = 4.24SGGGATTMTLEE954 pKa = 4.2SLVWAHH960 pKa = 6.54TGTSGAYY967 pKa = 9.04EE968 pKa = 3.96IYY970 pKa = 10.52KK971 pKa = 8.71NTVSGGDD978 pKa = 3.4GTVVVLIANTIDD990 pKa = 3.31VTLDD994 pKa = 2.94TFTAA998 pKa = 3.89
Molecular weight: 101.12 kDa
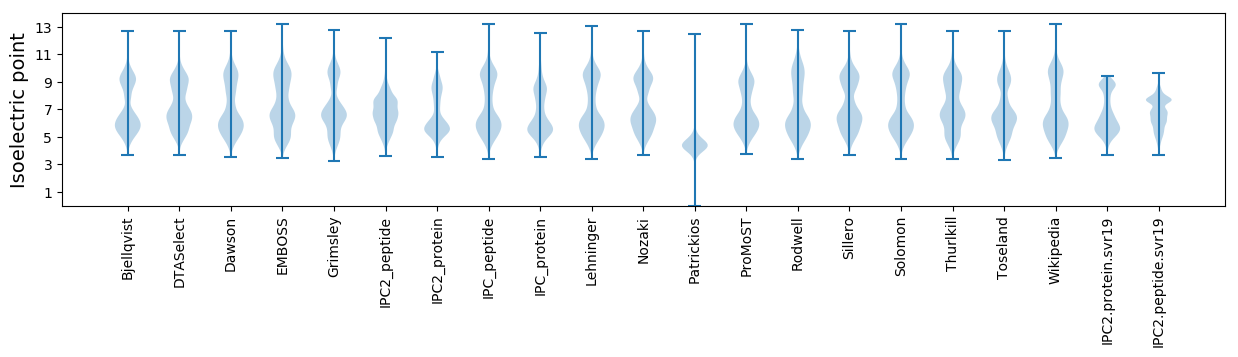
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8E1C6|A0A1H8E1C6_9BURK Cholesterol transport system auxiliary component OS=Duganella sp. CF402 OX=1855289 GN=SAMN05216319_5389 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84QVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84QVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1935185 |
33 |
5078 |
356.1 |
38.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.503 ± 0.054 | 0.794 ± 0.011 |
5.42 ± 0.03 | 5.102 ± 0.034 |
3.518 ± 0.021 | 8.063 ± 0.046 |
2.151 ± 0.017 | 4.828 ± 0.021 |
3.913 ± 0.03 | 10.47 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.015 | 3.309 ± 0.027 |
4.795 ± 0.027 | 4.217 ± 0.029 |
5.955 ± 0.034 | 5.791 ± 0.031 |
5.415 ± 0.039 | 7.231 ± 0.028 |
1.344 ± 0.015 | 2.684 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
