
Yellow-breasted capuchin simian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Simiispumavirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
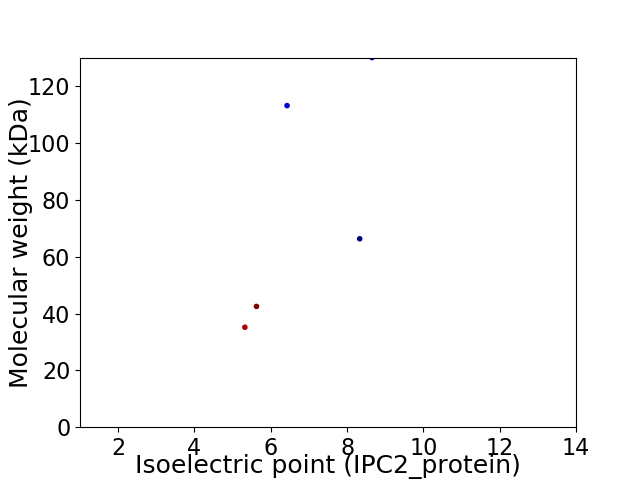
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
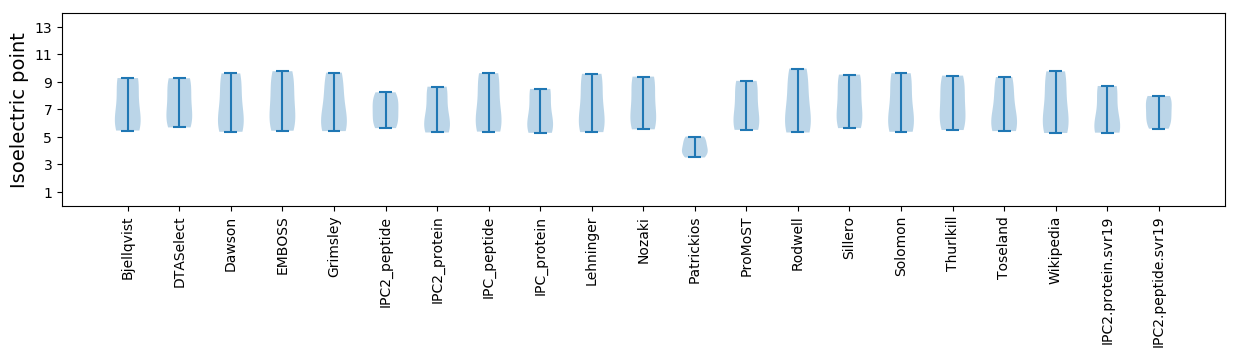
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3Y5C1|A0A0G3Y5C1_9RETR Env leader protein OS=Yellow-breasted capuchin simian foamy virus OX=2170206 GN=env PE=4 SV=1
MM1 pKa = 7.83AAQQEE6 pKa = 4.41EE7 pKa = 5.0TEE9 pKa = 4.33LKK11 pKa = 10.47EE12 pKa = 4.1FLAEE16 pKa = 4.85FPFNDD21 pKa = 4.31PDD23 pKa = 4.47DD24 pKa = 5.05DD25 pKa = 3.86LWVPVNIPPAPFQPYY40 pKa = 10.4ADD42 pKa = 3.64PHH44 pKa = 7.77DD45 pKa = 4.41EE46 pKa = 4.38EE47 pKa = 4.65EE48 pKa = 5.29GEE50 pKa = 4.17YY51 pKa = 10.77LLPKK55 pKa = 9.19EE56 pKa = 4.33QVFTTPSEE64 pKa = 4.37AEE66 pKa = 3.71TSEE69 pKa = 5.12RR70 pKa = 11.84DD71 pKa = 3.56DD72 pKa = 4.56PLPGTSTTINEE83 pKa = 4.36PTGEE87 pKa = 4.01EE88 pKa = 4.35PKK90 pKa = 10.9FKK92 pKa = 9.72WLQVARR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 3.95TDD102 pKa = 2.76YY103 pKa = 11.74DD104 pKa = 3.84RR105 pKa = 11.84FDD107 pKa = 3.83LGRR110 pKa = 11.84FFPDD114 pKa = 2.73RR115 pKa = 11.84LQRR118 pKa = 11.84EE119 pKa = 5.03FILQRR124 pKa = 11.84AILTAAGYY132 pKa = 10.05QPTLVEE138 pKa = 4.65HH139 pKa = 6.28CAKK142 pKa = 9.7MGWYY146 pKa = 10.23ACLQPHH152 pKa = 6.79HH153 pKa = 6.7GALGEE158 pKa = 4.29TTEE161 pKa = 4.86VYY163 pKa = 10.11YY164 pKa = 10.93KK165 pKa = 10.51CLKK168 pKa = 10.29CGSEE172 pKa = 3.79QWDD175 pKa = 3.66PLLYY179 pKa = 10.11IWDD182 pKa = 3.48KK183 pKa = 11.11HH184 pKa = 4.11MLMFKK189 pKa = 10.21RR190 pKa = 11.84AWTRR194 pKa = 11.84TPHH197 pKa = 5.69TSSPLSNLRR206 pKa = 11.84RR207 pKa = 11.84HH208 pKa = 6.6DD209 pKa = 4.91AICPGLNPLLPHH221 pKa = 6.93SSTEE225 pKa = 3.68PLLRR229 pKa = 11.84KK230 pKa = 9.19PRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.22PGDD237 pKa = 2.89RR238 pKa = 11.84WRR240 pKa = 11.84EE241 pKa = 3.74RR242 pKa = 11.84KK243 pKa = 9.56RR244 pKa = 11.84KK245 pKa = 8.95ISRR248 pKa = 11.84GGPTAVLPADD258 pKa = 3.64TFTYY262 pKa = 10.26ASEE265 pKa = 4.18WPQPEE270 pKa = 4.12RR271 pKa = 11.84MHH273 pKa = 7.21SGGLGTVPILVQTMQSKK290 pKa = 9.92CARR293 pKa = 11.84FEE295 pKa = 3.98MEE297 pKa = 3.83TLQNRR302 pKa = 11.84GIVV305 pKa = 3.3
MM1 pKa = 7.83AAQQEE6 pKa = 4.41EE7 pKa = 5.0TEE9 pKa = 4.33LKK11 pKa = 10.47EE12 pKa = 4.1FLAEE16 pKa = 4.85FPFNDD21 pKa = 4.31PDD23 pKa = 4.47DD24 pKa = 5.05DD25 pKa = 3.86LWVPVNIPPAPFQPYY40 pKa = 10.4ADD42 pKa = 3.64PHH44 pKa = 7.77DD45 pKa = 4.41EE46 pKa = 4.38EE47 pKa = 4.65EE48 pKa = 5.29GEE50 pKa = 4.17YY51 pKa = 10.77LLPKK55 pKa = 9.19EE56 pKa = 4.33QVFTTPSEE64 pKa = 4.37AEE66 pKa = 3.71TSEE69 pKa = 5.12RR70 pKa = 11.84DD71 pKa = 3.56DD72 pKa = 4.56PLPGTSTTINEE83 pKa = 4.36PTGEE87 pKa = 4.01EE88 pKa = 4.35PKK90 pKa = 10.9FKK92 pKa = 9.72WLQVARR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 3.95TDD102 pKa = 2.76YY103 pKa = 11.74DD104 pKa = 3.84RR105 pKa = 11.84FDD107 pKa = 3.83LGRR110 pKa = 11.84FFPDD114 pKa = 2.73RR115 pKa = 11.84LQRR118 pKa = 11.84EE119 pKa = 5.03FILQRR124 pKa = 11.84AILTAAGYY132 pKa = 10.05QPTLVEE138 pKa = 4.65HH139 pKa = 6.28CAKK142 pKa = 9.7MGWYY146 pKa = 10.23ACLQPHH152 pKa = 6.79HH153 pKa = 6.7GALGEE158 pKa = 4.29TTEE161 pKa = 4.86VYY163 pKa = 10.11YY164 pKa = 10.93KK165 pKa = 10.51CLKK168 pKa = 10.29CGSEE172 pKa = 3.79QWDD175 pKa = 3.66PLLYY179 pKa = 10.11IWDD182 pKa = 3.48KK183 pKa = 11.11HH184 pKa = 4.11MLMFKK189 pKa = 10.21RR190 pKa = 11.84AWTRR194 pKa = 11.84TPHH197 pKa = 5.69TSSPLSNLRR206 pKa = 11.84RR207 pKa = 11.84HH208 pKa = 6.6DD209 pKa = 4.91AICPGLNPLLPHH221 pKa = 6.93SSTEE225 pKa = 3.68PLLRR229 pKa = 11.84KK230 pKa = 9.19PRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.22PGDD237 pKa = 2.89RR238 pKa = 11.84WRR240 pKa = 11.84EE241 pKa = 3.74RR242 pKa = 11.84KK243 pKa = 9.56RR244 pKa = 11.84KK245 pKa = 8.95ISRR248 pKa = 11.84GGPTAVLPADD258 pKa = 3.64TFTYY262 pKa = 10.26ASEE265 pKa = 4.18WPQPEE270 pKa = 4.12RR271 pKa = 11.84MHH273 pKa = 7.21SGGLGTVPILVQTMQSKK290 pKa = 9.92CARR293 pKa = 11.84FEE295 pKa = 3.98MEE297 pKa = 3.83TLQNRR302 pKa = 11.84GIVV305 pKa = 3.3
Molecular weight: 35.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3Y3E1|A0A0G3Y3E1_9RETR Tas protein OS=Yellow-breasted capuchin simian foamy virus OX=2170206 GN=tas PE=4 SV=1
MM1 pKa = 7.28TAPPLLQLPVEE12 pKa = 4.51VKK14 pKa = 9.23KK15 pKa = 9.51TEE17 pKa = 4.54LNGFWDD23 pKa = 3.81TGAQITCIPEE33 pKa = 3.99AFLKK37 pKa = 10.8EE38 pKa = 4.14EE39 pKa = 4.1IPIGEE44 pKa = 4.11AQIKK48 pKa = 7.37TLHH51 pKa = 5.44GTKK54 pKa = 9.92LQPVYY59 pKa = 10.51YY60 pKa = 10.18LKK62 pKa = 10.9FKK64 pKa = 10.56ILGRR68 pKa = 11.84KK69 pKa = 8.75VEE71 pKa = 4.29AEE73 pKa = 4.16VTTSPFDD80 pKa = 3.54YY81 pKa = 11.04VIISPSDD88 pKa = 3.41IPWYY92 pKa = 10.27KK93 pKa = 10.0PQPLEE98 pKa = 4.12LTVKK102 pKa = 10.81LPVQDD107 pKa = 4.66FKK109 pKa = 11.8KK110 pKa = 10.75EE111 pKa = 4.23LINKK115 pKa = 9.37ANINNEE121 pKa = 4.14EE122 pKa = 4.11KK123 pKa = 10.66KK124 pKa = 10.53QLAKK128 pKa = 11.01LLDD131 pKa = 4.07KK132 pKa = 11.4YY133 pKa = 11.2DD134 pKa = 4.05ILWQQWEE141 pKa = 4.33NQVGHH146 pKa = 6.68RR147 pKa = 11.84KK148 pKa = 9.22IPPHH152 pKa = 6.48NIATGTVAPRR162 pKa = 11.84PQRR165 pKa = 11.84QYY167 pKa = 11.27HH168 pKa = 6.52INTKK172 pKa = 10.09AKK174 pKa = 10.19PSIQQVIDD182 pKa = 3.96DD183 pKa = 4.51LLKK186 pKa = 10.74QGVLVKK192 pKa = 9.25QTSVMNTPVYY202 pKa = 9.64PVPKK206 pKa = 9.66PDD208 pKa = 3.59GKK210 pKa = 9.91WRR212 pKa = 11.84MVLDD216 pKa = 3.44YY217 pKa = 10.78RR218 pKa = 11.84AVNKK222 pKa = 7.69TIPLIGAQNQHH233 pKa = 5.82SLGILTNLVRR243 pKa = 11.84QKK245 pKa = 10.91YY246 pKa = 9.88KK247 pKa = 10.25STIDD251 pKa = 3.45LSNGFWAHH259 pKa = 7.56PITKK263 pKa = 9.88DD264 pKa = 3.77SQWITAFTWEE274 pKa = 4.61GKK276 pKa = 6.41QHH278 pKa = 4.39VWTRR282 pKa = 11.84LPQGFLNSPALFTADD297 pKa = 4.02VVDD300 pKa = 4.66LLKK303 pKa = 10.56DD304 pKa = 3.19IPGISVYY311 pKa = 10.76VDD313 pKa = 4.41DD314 pKa = 5.79IYY316 pKa = 11.35FSTEE320 pKa = 3.79TVSEE324 pKa = 4.05HH325 pKa = 6.73LKK327 pKa = 10.38ILEE330 pKa = 4.2KK331 pKa = 10.65VFKK334 pKa = 10.28ILLEE338 pKa = 4.2AGYY341 pKa = 9.96IVSLKK346 pKa = 10.46KK347 pKa = 10.39SALLRR352 pKa = 11.84HH353 pKa = 5.81EE354 pKa = 4.26VTFLGFSITQTGRR367 pKa = 11.84GLTSEE372 pKa = 4.86FKK374 pKa = 11.01DD375 pKa = 3.35KK376 pKa = 10.64IQNITPPKK384 pKa = 8.33TLKK387 pKa = 10.15EE388 pKa = 4.02LQSILGLFNFARR400 pKa = 11.84NFVPNFSEE408 pKa = 4.44IIKK411 pKa = 9.68PLYY414 pKa = 10.46SLISTAEE421 pKa = 3.92GNNIKK426 pKa = 8.34WTSEE430 pKa = 3.75HH431 pKa = 5.72TRR433 pKa = 11.84HH434 pKa = 6.45LEE436 pKa = 4.31EE437 pKa = 4.54IVSALNHH444 pKa = 6.5AGNLEE449 pKa = 3.92QRR451 pKa = 11.84DD452 pKa = 4.16DD453 pKa = 3.97EE454 pKa = 4.72SPLVVKK460 pKa = 10.42LNASPKK466 pKa = 8.09TGYY469 pKa = 9.03IRR471 pKa = 11.84YY472 pKa = 8.85YY473 pKa = 10.79NKK475 pKa = 10.51GGQKK479 pKa = 9.37PIAYY483 pKa = 9.15ASHH486 pKa = 6.72VFTNTEE492 pKa = 4.05SKK494 pKa = 9.07FTPLEE499 pKa = 4.07KK500 pKa = 10.8LLVTMHH506 pKa = 7.05KK507 pKa = 10.73ALIKK511 pKa = 10.8AIDD514 pKa = 3.85LALGQPIEE522 pKa = 4.45VYY524 pKa = 10.75SPIVSMQKK532 pKa = 9.77LQKK535 pKa = 9.64TPLPEE540 pKa = 4.18RR541 pKa = 11.84KK542 pKa = 9.46ALSTRR547 pKa = 11.84WITWLSYY554 pKa = 11.25LEE556 pKa = 4.15DD557 pKa = 3.56PRR559 pKa = 11.84IIFHH563 pKa = 6.39YY564 pKa = 10.71DD565 pKa = 2.58KK566 pKa = 10.79TLPDD570 pKa = 4.04LKK572 pKa = 10.68NVPEE576 pKa = 4.86TITEE580 pKa = 4.13KK581 pKa = 10.46QPKK584 pKa = 8.45ILPIIEE590 pKa = 4.23YY591 pKa = 10.39AAVFYY596 pKa = 10.65TDD598 pKa = 3.37GSAIRR603 pKa = 11.84SPDD606 pKa = 3.12KK607 pKa = 11.19NKK609 pKa = 10.31SHH611 pKa = 6.66SSGMGIVQAIFKK623 pKa = 10.26PEE625 pKa = 3.77LTIEE629 pKa = 4.31HH630 pKa = 6.35QWTIPLGDD638 pKa = 3.58HH639 pKa = 5.95TAQYY643 pKa = 11.49AEE645 pKa = 3.83ISAVEE650 pKa = 4.21FACKK654 pKa = 9.69KK655 pKa = 10.2ANNISGPVLIVTDD668 pKa = 3.44SDD670 pKa = 4.12YY671 pKa = 11.43VARR674 pKa = 11.84SVNEE678 pKa = 3.8EE679 pKa = 3.95LPFWRR684 pKa = 11.84SNGFVNNKK692 pKa = 9.32KK693 pKa = 10.41KK694 pKa = 9.96PLKK697 pKa = 10.46HH698 pKa = 5.21ISKK701 pKa = 9.25WKK703 pKa = 10.21NISDD707 pKa = 3.87SLLLKK712 pKa = 10.21RR713 pKa = 11.84DD714 pKa = 3.11ITIVHH719 pKa = 6.41EE720 pKa = 4.73PGHH723 pKa = 5.75QPSHH727 pKa = 5.76TSIHH731 pKa = 4.92TQGNNLADD739 pKa = 3.7KK740 pKa = 10.66LATQGSYY747 pKa = 10.61NVNSIVKK754 pKa = 9.9NPSLDD759 pKa = 3.61AEE761 pKa = 4.47LEE763 pKa = 4.03QLINGHH769 pKa = 5.51SMKK772 pKa = 10.45GYY774 pKa = 7.63PSKK777 pKa = 11.0YY778 pKa = 10.15KK779 pKa = 10.85YY780 pKa = 9.77ILKK783 pKa = 9.88EE784 pKa = 3.71GQVFVLRR791 pKa = 11.84PEE793 pKa = 4.33GEE795 pKa = 4.05KK796 pKa = 10.11IIPPKK801 pKa = 10.06SDD803 pKa = 2.8RR804 pKa = 11.84PALVKK809 pKa = 10.18VAHH812 pKa = 6.42EE813 pKa = 4.39FSHH816 pKa = 6.99AGRR819 pKa = 11.84EE820 pKa = 4.33ATVLRR825 pKa = 11.84LQDD828 pKa = 3.73KK829 pKa = 9.13YY830 pKa = 9.53WWPNMRR836 pKa = 11.84KK837 pKa = 9.73DD838 pKa = 4.01VISHH842 pKa = 5.81IRR844 pKa = 11.84MCKK847 pKa = 9.43PCLTTDD853 pKa = 3.45SSNLTPIPPKK863 pKa = 9.62PQQRR867 pKa = 11.84PKK869 pKa = 10.99KK870 pKa = 9.28PFDD873 pKa = 3.57KK874 pKa = 11.01FFIDD878 pKa = 4.29YY879 pKa = 10.24IGPLPPSHH887 pKa = 6.3GFAYY891 pKa = 10.62VLVVVDD897 pKa = 4.38AATGFTWLYY906 pKa = 5.84PTKK909 pKa = 10.62APSTNATITSLNILLGTAVPRR930 pKa = 11.84VLHH933 pKa = 6.51SDD935 pKa = 2.92QGSAFTSSAFADD947 pKa = 3.46WAKK950 pKa = 10.97EE951 pKa = 3.67KK952 pKa = 10.79GIQLEE957 pKa = 4.54FSTPYY962 pKa = 10.11HH963 pKa = 5.86PQSSGMVEE971 pKa = 3.9RR972 pKa = 11.84KK973 pKa = 9.31NRR975 pKa = 11.84EE976 pKa = 3.56IKK978 pKa = 10.6RR979 pKa = 11.84LITKK983 pKa = 9.73LLVGRR988 pKa = 11.84PTKK991 pKa = 9.9WYY993 pKa = 9.08PLIPTIQLALNNTYY1007 pKa = 10.43SVHH1010 pKa = 5.57YY1011 pKa = 9.93KK1012 pKa = 8.78KK1013 pKa = 9.09TPHH1016 pKa = 5.97QLLFGVDD1023 pKa = 3.65GNVPFANQDD1032 pKa = 3.55TLDD1035 pKa = 3.66LTRR1038 pKa = 11.84EE1039 pKa = 4.19EE1040 pKa = 4.36EE1041 pKa = 4.11LSLLSEE1047 pKa = 4.5VRR1049 pKa = 11.84TSLLPPSTPPASRR1062 pKa = 11.84RR1063 pKa = 11.84SWLPSVGLLVQEE1075 pKa = 4.63RR1076 pKa = 11.84VARR1079 pKa = 11.84PSQLRR1084 pKa = 11.84PKK1086 pKa = 8.41WRR1088 pKa = 11.84KK1089 pKa = 5.65PTPILEE1095 pKa = 4.4VVNDD1099 pKa = 3.9RR1100 pKa = 11.84TVVILDD1106 pKa = 3.4NQGQRR1111 pKa = 11.84RR1112 pKa = 11.84TVSIDD1117 pKa = 3.24NLKK1120 pKa = 8.92LTPHH1124 pKa = 6.49QDD1126 pKa = 3.12GTSNEE1131 pKa = 4.16PNGVDD1136 pKa = 3.34PLEE1139 pKa = 4.1QEE1141 pKa = 4.51KK1142 pKa = 10.3EE1143 pKa = 4.02HH1144 pKa = 7.19DD1145 pKa = 3.55NHH1147 pKa = 7.62DD1148 pKa = 3.37II1149 pKa = 4.09
MM1 pKa = 7.28TAPPLLQLPVEE12 pKa = 4.51VKK14 pKa = 9.23KK15 pKa = 9.51TEE17 pKa = 4.54LNGFWDD23 pKa = 3.81TGAQITCIPEE33 pKa = 3.99AFLKK37 pKa = 10.8EE38 pKa = 4.14EE39 pKa = 4.1IPIGEE44 pKa = 4.11AQIKK48 pKa = 7.37TLHH51 pKa = 5.44GTKK54 pKa = 9.92LQPVYY59 pKa = 10.51YY60 pKa = 10.18LKK62 pKa = 10.9FKK64 pKa = 10.56ILGRR68 pKa = 11.84KK69 pKa = 8.75VEE71 pKa = 4.29AEE73 pKa = 4.16VTTSPFDD80 pKa = 3.54YY81 pKa = 11.04VIISPSDD88 pKa = 3.41IPWYY92 pKa = 10.27KK93 pKa = 10.0PQPLEE98 pKa = 4.12LTVKK102 pKa = 10.81LPVQDD107 pKa = 4.66FKK109 pKa = 11.8KK110 pKa = 10.75EE111 pKa = 4.23LINKK115 pKa = 9.37ANINNEE121 pKa = 4.14EE122 pKa = 4.11KK123 pKa = 10.66KK124 pKa = 10.53QLAKK128 pKa = 11.01LLDD131 pKa = 4.07KK132 pKa = 11.4YY133 pKa = 11.2DD134 pKa = 4.05ILWQQWEE141 pKa = 4.33NQVGHH146 pKa = 6.68RR147 pKa = 11.84KK148 pKa = 9.22IPPHH152 pKa = 6.48NIATGTVAPRR162 pKa = 11.84PQRR165 pKa = 11.84QYY167 pKa = 11.27HH168 pKa = 6.52INTKK172 pKa = 10.09AKK174 pKa = 10.19PSIQQVIDD182 pKa = 3.96DD183 pKa = 4.51LLKK186 pKa = 10.74QGVLVKK192 pKa = 9.25QTSVMNTPVYY202 pKa = 9.64PVPKK206 pKa = 9.66PDD208 pKa = 3.59GKK210 pKa = 9.91WRR212 pKa = 11.84MVLDD216 pKa = 3.44YY217 pKa = 10.78RR218 pKa = 11.84AVNKK222 pKa = 7.69TIPLIGAQNQHH233 pKa = 5.82SLGILTNLVRR243 pKa = 11.84QKK245 pKa = 10.91YY246 pKa = 9.88KK247 pKa = 10.25STIDD251 pKa = 3.45LSNGFWAHH259 pKa = 7.56PITKK263 pKa = 9.88DD264 pKa = 3.77SQWITAFTWEE274 pKa = 4.61GKK276 pKa = 6.41QHH278 pKa = 4.39VWTRR282 pKa = 11.84LPQGFLNSPALFTADD297 pKa = 4.02VVDD300 pKa = 4.66LLKK303 pKa = 10.56DD304 pKa = 3.19IPGISVYY311 pKa = 10.76VDD313 pKa = 4.41DD314 pKa = 5.79IYY316 pKa = 11.35FSTEE320 pKa = 3.79TVSEE324 pKa = 4.05HH325 pKa = 6.73LKK327 pKa = 10.38ILEE330 pKa = 4.2KK331 pKa = 10.65VFKK334 pKa = 10.28ILLEE338 pKa = 4.2AGYY341 pKa = 9.96IVSLKK346 pKa = 10.46KK347 pKa = 10.39SALLRR352 pKa = 11.84HH353 pKa = 5.81EE354 pKa = 4.26VTFLGFSITQTGRR367 pKa = 11.84GLTSEE372 pKa = 4.86FKK374 pKa = 11.01DD375 pKa = 3.35KK376 pKa = 10.64IQNITPPKK384 pKa = 8.33TLKK387 pKa = 10.15EE388 pKa = 4.02LQSILGLFNFARR400 pKa = 11.84NFVPNFSEE408 pKa = 4.44IIKK411 pKa = 9.68PLYY414 pKa = 10.46SLISTAEE421 pKa = 3.92GNNIKK426 pKa = 8.34WTSEE430 pKa = 3.75HH431 pKa = 5.72TRR433 pKa = 11.84HH434 pKa = 6.45LEE436 pKa = 4.31EE437 pKa = 4.54IVSALNHH444 pKa = 6.5AGNLEE449 pKa = 3.92QRR451 pKa = 11.84DD452 pKa = 4.16DD453 pKa = 3.97EE454 pKa = 4.72SPLVVKK460 pKa = 10.42LNASPKK466 pKa = 8.09TGYY469 pKa = 9.03IRR471 pKa = 11.84YY472 pKa = 8.85YY473 pKa = 10.79NKK475 pKa = 10.51GGQKK479 pKa = 9.37PIAYY483 pKa = 9.15ASHH486 pKa = 6.72VFTNTEE492 pKa = 4.05SKK494 pKa = 9.07FTPLEE499 pKa = 4.07KK500 pKa = 10.8LLVTMHH506 pKa = 7.05KK507 pKa = 10.73ALIKK511 pKa = 10.8AIDD514 pKa = 3.85LALGQPIEE522 pKa = 4.45VYY524 pKa = 10.75SPIVSMQKK532 pKa = 9.77LQKK535 pKa = 9.64TPLPEE540 pKa = 4.18RR541 pKa = 11.84KK542 pKa = 9.46ALSTRR547 pKa = 11.84WITWLSYY554 pKa = 11.25LEE556 pKa = 4.15DD557 pKa = 3.56PRR559 pKa = 11.84IIFHH563 pKa = 6.39YY564 pKa = 10.71DD565 pKa = 2.58KK566 pKa = 10.79TLPDD570 pKa = 4.04LKK572 pKa = 10.68NVPEE576 pKa = 4.86TITEE580 pKa = 4.13KK581 pKa = 10.46QPKK584 pKa = 8.45ILPIIEE590 pKa = 4.23YY591 pKa = 10.39AAVFYY596 pKa = 10.65TDD598 pKa = 3.37GSAIRR603 pKa = 11.84SPDD606 pKa = 3.12KK607 pKa = 11.19NKK609 pKa = 10.31SHH611 pKa = 6.66SSGMGIVQAIFKK623 pKa = 10.26PEE625 pKa = 3.77LTIEE629 pKa = 4.31HH630 pKa = 6.35QWTIPLGDD638 pKa = 3.58HH639 pKa = 5.95TAQYY643 pKa = 11.49AEE645 pKa = 3.83ISAVEE650 pKa = 4.21FACKK654 pKa = 9.69KK655 pKa = 10.2ANNISGPVLIVTDD668 pKa = 3.44SDD670 pKa = 4.12YY671 pKa = 11.43VARR674 pKa = 11.84SVNEE678 pKa = 3.8EE679 pKa = 3.95LPFWRR684 pKa = 11.84SNGFVNNKK692 pKa = 9.32KK693 pKa = 10.41KK694 pKa = 9.96PLKK697 pKa = 10.46HH698 pKa = 5.21ISKK701 pKa = 9.25WKK703 pKa = 10.21NISDD707 pKa = 3.87SLLLKK712 pKa = 10.21RR713 pKa = 11.84DD714 pKa = 3.11ITIVHH719 pKa = 6.41EE720 pKa = 4.73PGHH723 pKa = 5.75QPSHH727 pKa = 5.76TSIHH731 pKa = 4.92TQGNNLADD739 pKa = 3.7KK740 pKa = 10.66LATQGSYY747 pKa = 10.61NVNSIVKK754 pKa = 9.9NPSLDD759 pKa = 3.61AEE761 pKa = 4.47LEE763 pKa = 4.03QLINGHH769 pKa = 5.51SMKK772 pKa = 10.45GYY774 pKa = 7.63PSKK777 pKa = 11.0YY778 pKa = 10.15KK779 pKa = 10.85YY780 pKa = 9.77ILKK783 pKa = 9.88EE784 pKa = 3.71GQVFVLRR791 pKa = 11.84PEE793 pKa = 4.33GEE795 pKa = 4.05KK796 pKa = 10.11IIPPKK801 pKa = 10.06SDD803 pKa = 2.8RR804 pKa = 11.84PALVKK809 pKa = 10.18VAHH812 pKa = 6.42EE813 pKa = 4.39FSHH816 pKa = 6.99AGRR819 pKa = 11.84EE820 pKa = 4.33ATVLRR825 pKa = 11.84LQDD828 pKa = 3.73KK829 pKa = 9.13YY830 pKa = 9.53WWPNMRR836 pKa = 11.84KK837 pKa = 9.73DD838 pKa = 4.01VISHH842 pKa = 5.81IRR844 pKa = 11.84MCKK847 pKa = 9.43PCLTTDD853 pKa = 3.45SSNLTPIPPKK863 pKa = 9.62PQQRR867 pKa = 11.84PKK869 pKa = 10.99KK870 pKa = 9.28PFDD873 pKa = 3.57KK874 pKa = 11.01FFIDD878 pKa = 4.29YY879 pKa = 10.24IGPLPPSHH887 pKa = 6.3GFAYY891 pKa = 10.62VLVVVDD897 pKa = 4.38AATGFTWLYY906 pKa = 5.84PTKK909 pKa = 10.62APSTNATITSLNILLGTAVPRR930 pKa = 11.84VLHH933 pKa = 6.51SDD935 pKa = 2.92QGSAFTSSAFADD947 pKa = 3.46WAKK950 pKa = 10.97EE951 pKa = 3.67KK952 pKa = 10.79GIQLEE957 pKa = 4.54FSTPYY962 pKa = 10.11HH963 pKa = 5.86PQSSGMVEE971 pKa = 3.9RR972 pKa = 11.84KK973 pKa = 9.31NRR975 pKa = 11.84EE976 pKa = 3.56IKK978 pKa = 10.6RR979 pKa = 11.84LITKK983 pKa = 9.73LLVGRR988 pKa = 11.84PTKK991 pKa = 9.9WYY993 pKa = 9.08PLIPTIQLALNNTYY1007 pKa = 10.43SVHH1010 pKa = 5.57YY1011 pKa = 9.93KK1012 pKa = 8.78KK1013 pKa = 9.09TPHH1016 pKa = 5.97QLLFGVDD1023 pKa = 3.65GNVPFANQDD1032 pKa = 3.55TLDD1035 pKa = 3.66LTRR1038 pKa = 11.84EE1039 pKa = 4.19EE1040 pKa = 4.36EE1041 pKa = 4.11LSLLSEE1047 pKa = 4.5VRR1049 pKa = 11.84TSLLPPSTPPASRR1062 pKa = 11.84RR1063 pKa = 11.84SWLPSVGLLVQEE1075 pKa = 4.63RR1076 pKa = 11.84VARR1079 pKa = 11.84PSQLRR1084 pKa = 11.84PKK1086 pKa = 8.41WRR1088 pKa = 11.84KK1089 pKa = 5.65PTPILEE1095 pKa = 4.4VVNDD1099 pKa = 3.9RR1100 pKa = 11.84TVVILDD1106 pKa = 3.4NQGQRR1111 pKa = 11.84RR1112 pKa = 11.84TVSIDD1117 pKa = 3.24NLKK1120 pKa = 8.92LTPHH1124 pKa = 6.49QDD1126 pKa = 3.12GTSNEE1131 pKa = 4.16PNGVDD1136 pKa = 3.34PLEE1139 pKa = 4.1QEE1141 pKa = 4.51KK1142 pKa = 10.3EE1143 pKa = 4.02HH1144 pKa = 7.19DD1145 pKa = 3.55NHH1147 pKa = 7.62DD1148 pKa = 3.37II1149 pKa = 4.09
Molecular weight: 130.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3429 |
305 |
1149 |
685.8 |
77.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.628 ± 0.529 | 1.633 ± 0.626 |
4.608 ± 0.168 | 5.862 ± 0.576 |
3.12 ± 0.182 | 5.745 ± 0.927 |
2.45 ± 0.324 | 6.183 ± 1.073 |
5.92 ± 1.295 | 9.536 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.604 ± 0.427 | 5.133 ± 0.55 |
8.107 ± 1.17 | 5.016 ± 0.303 |
4.958 ± 0.748 | 6.328 ± 0.688 |
6.737 ± 0.325 | 5.862 ± 0.367 |
2.071 ± 0.24 | 3.5 ± 0.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
