
Sweet potato leaf curl Hubei virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 7.81
Get precalculated fractions of proteins
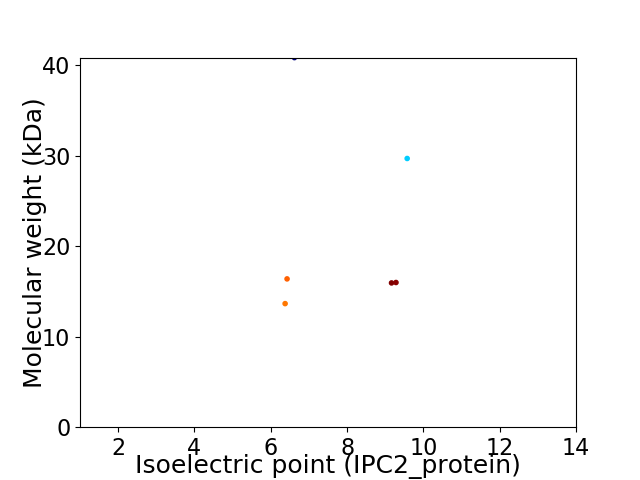
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345GSG2|A0A345GSG2_9GEMI Protein V2 OS=Sweet potato leaf curl Hubei virus OX=2282485 PE=3 SV=1
MM1 pKa = 7.17EE2 pKa = 4.15QSLWDD7 pKa = 4.38PLTHH11 pKa = 6.93PLPEE15 pKa = 4.21TLHH18 pKa = 6.51GFRR21 pKa = 11.84CMLSLKK27 pKa = 10.18YY28 pKa = 10.11MSNIRR33 pKa = 11.84DD34 pKa = 3.47KK35 pKa = 11.56YY36 pKa = 9.73EE37 pKa = 4.39PGTLGHH43 pKa = 6.61EE44 pKa = 4.23LAVLLIRR51 pKa = 11.84SLRR54 pKa = 11.84GKK56 pKa = 10.16NYY58 pKa = 9.19VRR60 pKa = 11.84STSSYY65 pKa = 10.99EE66 pKa = 3.99EE67 pKa = 4.04VCALLSEE74 pKa = 4.68TAGSSPSKK82 pKa = 10.03LHH84 pKa = 6.56DD85 pKa = 3.16RR86 pKa = 11.84HH87 pKa = 6.5RR88 pKa = 11.84PVCGEE93 pKa = 4.33CCPACCSNVRR103 pKa = 11.84SSSCEE108 pKa = 3.54GKK110 pKa = 10.29EE111 pKa = 3.64ADD113 pKa = 3.54LSKK116 pKa = 10.75EE117 pKa = 3.77RR118 pKa = 11.84RR119 pKa = 11.84LDD121 pKa = 3.48SS122 pKa = 3.79
MM1 pKa = 7.17EE2 pKa = 4.15QSLWDD7 pKa = 4.38PLTHH11 pKa = 6.93PLPEE15 pKa = 4.21TLHH18 pKa = 6.51GFRR21 pKa = 11.84CMLSLKK27 pKa = 10.18YY28 pKa = 10.11MSNIRR33 pKa = 11.84DD34 pKa = 3.47KK35 pKa = 11.56YY36 pKa = 9.73EE37 pKa = 4.39PGTLGHH43 pKa = 6.61EE44 pKa = 4.23LAVLLIRR51 pKa = 11.84SLRR54 pKa = 11.84GKK56 pKa = 10.16NYY58 pKa = 9.19VRR60 pKa = 11.84STSSYY65 pKa = 10.99EE66 pKa = 3.99EE67 pKa = 4.04VCALLSEE74 pKa = 4.68TAGSSPSKK82 pKa = 10.03LHH84 pKa = 6.56DD85 pKa = 3.16RR86 pKa = 11.84HH87 pKa = 6.5RR88 pKa = 11.84PVCGEE93 pKa = 4.33CCPACCSNVRR103 pKa = 11.84SSSCEE108 pKa = 3.54GKK110 pKa = 10.29EE111 pKa = 3.64ADD113 pKa = 3.54LSKK116 pKa = 10.75EE117 pKa = 3.77RR118 pKa = 11.84RR119 pKa = 11.84LDD121 pKa = 3.48SS122 pKa = 3.79
Molecular weight: 13.66 kDa
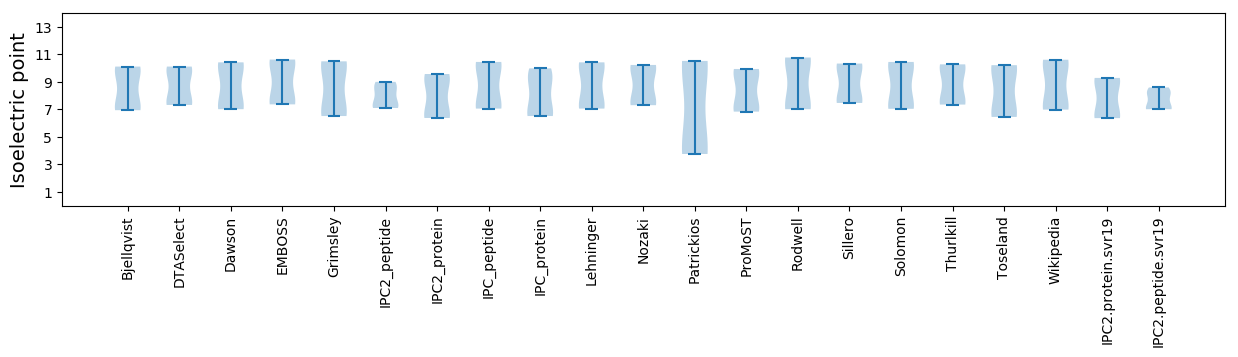
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345GSF9|A0A345GSF9_9GEMI AC4 OS=Sweet potato leaf curl Hubei virus OX=2282485 PE=3 SV=1
MM1 pKa = 7.79SDD3 pKa = 3.26RR4 pKa = 11.84LRR6 pKa = 11.84VTRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.01APYY14 pKa = 10.32SRR16 pKa = 11.84RR17 pKa = 11.84PQAARR22 pKa = 11.84RR23 pKa = 11.84LNFTTDD29 pKa = 2.51IVPYY33 pKa = 9.65VGNAAPLAAATYY45 pKa = 10.31VPVPVKK51 pKa = 10.37ARR53 pKa = 11.84KK54 pKa = 7.52RR55 pKa = 11.84TSRR58 pKa = 11.84KK59 pKa = 9.25RR60 pKa = 11.84GDD62 pKa = 3.66WIPRR66 pKa = 11.84GCVGPCKK73 pKa = 10.08VQDD76 pKa = 3.86YY77 pKa = 8.72EE78 pKa = 4.55FKK80 pKa = 10.3MDD82 pKa = 3.6VPHH85 pKa = 7.17GGTFVCVSDD94 pKa = 3.96FTRR97 pKa = 11.84GTGLTHH103 pKa = 7.28RR104 pKa = 11.84LGKK107 pKa = 9.86RR108 pKa = 11.84VCIKK112 pKa = 11.1SMGFSGKK119 pKa = 7.94VWMDD123 pKa = 3.53DD124 pKa = 3.61NIAKK128 pKa = 9.84KK129 pKa = 10.52DD130 pKa = 3.43HH131 pKa = 6.37TNIITFWLVRR141 pKa = 11.84DD142 pKa = 3.76RR143 pKa = 11.84RR144 pKa = 11.84PNKK147 pKa = 10.42DD148 pKa = 3.21PLTFSQLFHH157 pKa = 6.56MFDD160 pKa = 4.05NEE162 pKa = 3.9PLTAKK167 pKa = 10.05VRR169 pKa = 11.84TDD171 pKa = 2.9LRR173 pKa = 11.84DD174 pKa = 3.17RR175 pKa = 11.84FQVLRR180 pKa = 11.84TFSVTVSGGPYY191 pKa = 9.23AHH193 pKa = 7.66KK194 pKa = 10.09EE195 pKa = 3.71QAQVRR200 pKa = 11.84RR201 pKa = 11.84FFKK204 pKa = 10.71GLNNHH209 pKa = 6.26VIYY212 pKa = 10.49NHH214 pKa = 6.53KK215 pKa = 10.73EE216 pKa = 3.41EE217 pKa = 4.8AKK219 pKa = 10.88YY220 pKa = 10.75EE221 pKa = 4.12NQLEE225 pKa = 4.18NAMLVYY231 pKa = 10.27SASSHH236 pKa = 6.1ASNPVYY242 pKa = 10.32QSLRR246 pKa = 11.84VRR248 pKa = 11.84AYY250 pKa = 10.48FYY252 pKa = 10.58DD253 pKa = 3.16SHH255 pKa = 6.48MNN257 pKa = 3.42
MM1 pKa = 7.79SDD3 pKa = 3.26RR4 pKa = 11.84LRR6 pKa = 11.84VTRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.01APYY14 pKa = 10.32SRR16 pKa = 11.84RR17 pKa = 11.84PQAARR22 pKa = 11.84RR23 pKa = 11.84LNFTTDD29 pKa = 2.51IVPYY33 pKa = 9.65VGNAAPLAAATYY45 pKa = 10.31VPVPVKK51 pKa = 10.37ARR53 pKa = 11.84KK54 pKa = 7.52RR55 pKa = 11.84TSRR58 pKa = 11.84KK59 pKa = 9.25RR60 pKa = 11.84GDD62 pKa = 3.66WIPRR66 pKa = 11.84GCVGPCKK73 pKa = 10.08VQDD76 pKa = 3.86YY77 pKa = 8.72EE78 pKa = 4.55FKK80 pKa = 10.3MDD82 pKa = 3.6VPHH85 pKa = 7.17GGTFVCVSDD94 pKa = 3.96FTRR97 pKa = 11.84GTGLTHH103 pKa = 7.28RR104 pKa = 11.84LGKK107 pKa = 9.86RR108 pKa = 11.84VCIKK112 pKa = 11.1SMGFSGKK119 pKa = 7.94VWMDD123 pKa = 3.53DD124 pKa = 3.61NIAKK128 pKa = 9.84KK129 pKa = 10.52DD130 pKa = 3.43HH131 pKa = 6.37TNIITFWLVRR141 pKa = 11.84DD142 pKa = 3.76RR143 pKa = 11.84RR144 pKa = 11.84PNKK147 pKa = 10.42DD148 pKa = 3.21PLTFSQLFHH157 pKa = 6.56MFDD160 pKa = 4.05NEE162 pKa = 3.9PLTAKK167 pKa = 10.05VRR169 pKa = 11.84TDD171 pKa = 2.9LRR173 pKa = 11.84DD174 pKa = 3.17RR175 pKa = 11.84FQVLRR180 pKa = 11.84TFSVTVSGGPYY191 pKa = 9.23AHH193 pKa = 7.66KK194 pKa = 10.09EE195 pKa = 3.71QAQVRR200 pKa = 11.84RR201 pKa = 11.84FFKK204 pKa = 10.71GLNNHH209 pKa = 6.26VIYY212 pKa = 10.49NHH214 pKa = 6.53KK215 pKa = 10.73EE216 pKa = 3.41EE217 pKa = 4.8AKK219 pKa = 10.88YY220 pKa = 10.75EE221 pKa = 4.12NQLEE225 pKa = 4.18NAMLVYY231 pKa = 10.27SASSHH236 pKa = 6.1ASNPVYY242 pKa = 10.32QSLRR246 pKa = 11.84VRR248 pKa = 11.84AYY250 pKa = 10.48FYY252 pKa = 10.58DD253 pKa = 3.16SHH255 pKa = 6.48MNN257 pKa = 3.42
Molecular weight: 29.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165 |
122 |
360 |
194.2 |
22.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.408 ± 0.805 | 2.661 ± 0.861 |
4.807 ± 0.412 | 4.979 ± 1.091 |
4.206 ± 0.581 | 5.236 ± 0.278 |
3.691 ± 0.346 | 5.15 ± 1.054 |
5.923 ± 0.395 | 8.24 ± 1.262 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.608 | 4.549 ± 0.883 |
6.438 ± 0.748 | 4.378 ± 0.66 |
6.867 ± 1.138 | 9.871 ± 1.614 |
5.408 ± 0.851 | 5.15 ± 1.032 |
1.459 ± 0.214 | 3.605 ± 0.49 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
