
Capybara microvirus Cap1_SP_131
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 4.94
Get precalculated fractions of proteins
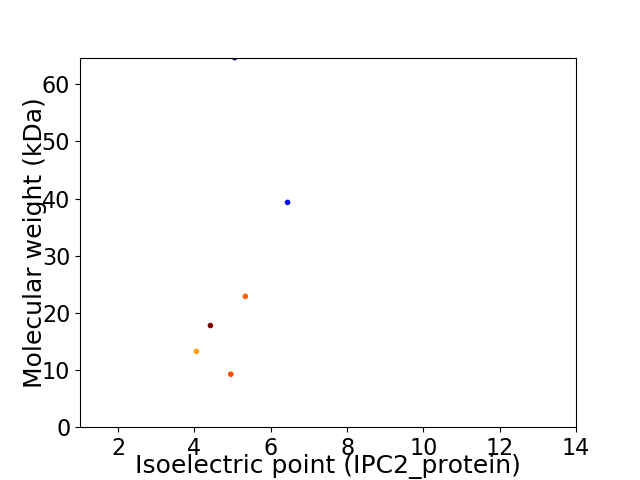
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
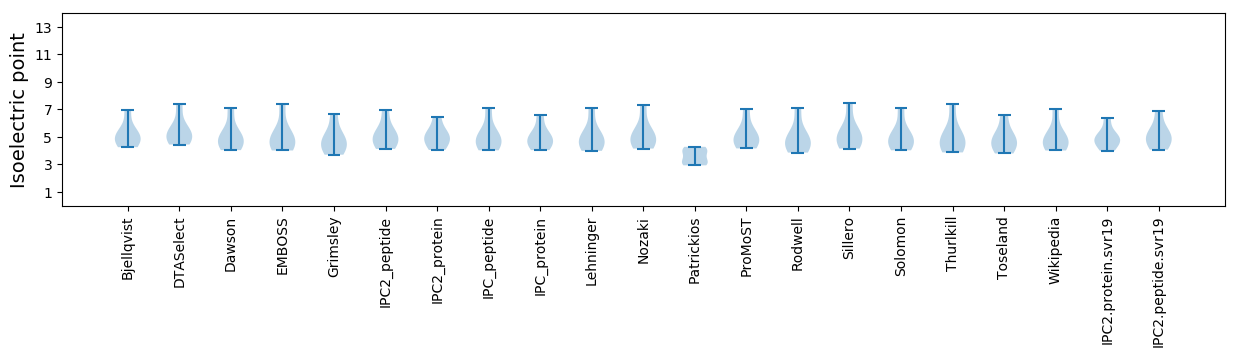
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7U5|A0A4P8W7U5_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_131 OX=2585382 PE=4 SV=1
MM1 pKa = 7.76FDD3 pKa = 4.1VDD5 pKa = 4.45DD6 pKa = 5.07LNIFRR11 pKa = 11.84PFFHH15 pKa = 7.56DD16 pKa = 5.89AILEE20 pKa = 4.26SSGDD24 pKa = 3.67VYY26 pKa = 11.31SWSDD30 pKa = 3.47LSLGAPLLPDD40 pKa = 4.36TILNAVCFKK49 pKa = 10.67VSEE52 pKa = 4.2RR53 pKa = 11.84LVSRR57 pKa = 11.84FLMLLRR63 pKa = 11.84SGFNCYY69 pKa = 9.88FDD71 pKa = 4.54LSYY74 pKa = 11.36DD75 pKa = 3.98FSPARR80 pKa = 11.84DD81 pKa = 3.63DD82 pKa = 4.23FDD84 pKa = 4.59FFFMVEE90 pKa = 4.17PLCADD95 pKa = 3.44DD96 pKa = 5.66CDD98 pKa = 5.24IVLSWLIKK106 pKa = 9.8YY107 pKa = 9.57CFLKK111 pKa = 10.98VKK113 pKa = 9.97EE114 pKa = 4.12
MM1 pKa = 7.76FDD3 pKa = 4.1VDD5 pKa = 4.45DD6 pKa = 5.07LNIFRR11 pKa = 11.84PFFHH15 pKa = 7.56DD16 pKa = 5.89AILEE20 pKa = 4.26SSGDD24 pKa = 3.67VYY26 pKa = 11.31SWSDD30 pKa = 3.47LSLGAPLLPDD40 pKa = 4.36TILNAVCFKK49 pKa = 10.67VSEE52 pKa = 4.2RR53 pKa = 11.84LVSRR57 pKa = 11.84FLMLLRR63 pKa = 11.84SGFNCYY69 pKa = 9.88FDD71 pKa = 4.54LSYY74 pKa = 11.36DD75 pKa = 3.98FSPARR80 pKa = 11.84DD81 pKa = 3.63DD82 pKa = 4.23FDD84 pKa = 4.59FFFMVEE90 pKa = 4.17PLCADD95 pKa = 3.44DD96 pKa = 5.66CDD98 pKa = 5.24IVLSWLIKK106 pKa = 9.8YY107 pKa = 9.57CFLKK111 pKa = 10.98VKK113 pKa = 9.97EE114 pKa = 4.12
Molecular weight: 13.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W481|A0A4P8W481_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_131 OX=2585382 PE=3 SV=1
MM1 pKa = 7.04FTTFKK6 pKa = 10.61KK7 pKa = 10.52DD8 pKa = 3.01EE9 pKa = 4.22YY10 pKa = 11.02DD11 pKa = 3.67FLVPEE16 pKa = 4.1YY17 pKa = 10.68AFNRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84QLFPRR28 pKa = 11.84ADD30 pKa = 3.65LPGLVNPIDD39 pKa = 4.08YY40 pKa = 10.35EE41 pKa = 4.09NSQLDD46 pKa = 3.44LSIPCRR52 pKa = 11.84TCIGCRR58 pKa = 11.84LDD60 pKa = 5.38QSRR63 pKa = 11.84QWADD67 pKa = 2.63RR68 pKa = 11.84MMIEE72 pKa = 5.16LDD74 pKa = 3.3HH75 pKa = 6.63SKK77 pKa = 10.95CGIFVTLTYY86 pKa = 10.75DD87 pKa = 3.19PAFAHH92 pKa = 6.76PCSIDD97 pKa = 3.27SDD99 pKa = 3.8DD100 pKa = 3.59RR101 pKa = 11.84VYY103 pKa = 11.56YY104 pKa = 10.75SLDD107 pKa = 3.31VRR109 pKa = 11.84DD110 pKa = 3.63TQLFNKK116 pKa = 9.26SLRR119 pKa = 11.84KK120 pKa = 9.82AFPDD124 pKa = 3.07RR125 pKa = 11.84KK126 pKa = 9.97IRR128 pKa = 11.84FFLAGEE134 pKa = 4.22YY135 pKa = 10.81GPSTWRR141 pKa = 11.84PHH143 pKa = 3.84YY144 pKa = 9.6HH145 pKa = 6.07AVYY148 pKa = 10.06FGLSIDD154 pKa = 4.72DD155 pKa = 5.26FPDD158 pKa = 3.76LMPLKK163 pKa = 9.2RR164 pKa = 11.84TSAGSLIYY172 pKa = 10.23TSEE175 pKa = 4.15KK176 pKa = 10.18LQKK179 pKa = 8.78LWKK182 pKa = 9.88RR183 pKa = 11.84GFISLSPVSWQTCAYY198 pKa = 8.04VARR201 pKa = 11.84YY202 pKa = 5.27TTKK205 pKa = 10.96KK206 pKa = 10.74LIDD209 pKa = 4.27SNPLMSDD216 pKa = 3.0YY217 pKa = 11.48DD218 pKa = 4.4LFNMHH223 pKa = 7.43PEE225 pKa = 3.91FMTCSRR231 pKa = 11.84KK232 pKa = 9.69PGLSGYY238 pKa = 10.31FYY240 pKa = 10.56DD241 pKa = 4.79DD242 pKa = 4.41HH243 pKa = 7.5PDD245 pKa = 3.63LVPDD249 pKa = 4.94SIDD252 pKa = 3.3DD253 pKa = 3.7FKK255 pKa = 11.36IYY257 pKa = 10.27IRR259 pKa = 11.84DD260 pKa = 3.68PDD262 pKa = 3.56GLKK265 pKa = 10.08PVKK268 pKa = 10.09SVKK271 pKa = 8.75TPKK274 pKa = 10.5YY275 pKa = 10.33LLKK278 pKa = 10.37QLQAVNPLLYY288 pKa = 10.51NEE290 pKa = 4.3YY291 pKa = 10.63VDD293 pKa = 3.7MQRR296 pKa = 11.84VSSEE300 pKa = 3.99DD301 pKa = 3.39SLLLKK306 pKa = 10.2LQRR309 pKa = 11.84TSLPEE314 pKa = 3.59AEE316 pKa = 4.78YY317 pKa = 10.51ILSEE321 pKa = 4.33EE322 pKa = 4.17FTKK325 pKa = 10.91QNSAKK330 pKa = 10.45KK331 pKa = 10.45LIRR334 pKa = 11.84SDD336 pKa = 3.5LL337 pKa = 3.76
MM1 pKa = 7.04FTTFKK6 pKa = 10.61KK7 pKa = 10.52DD8 pKa = 3.01EE9 pKa = 4.22YY10 pKa = 11.02DD11 pKa = 3.67FLVPEE16 pKa = 4.1YY17 pKa = 10.68AFNRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84QLFPRR28 pKa = 11.84ADD30 pKa = 3.65LPGLVNPIDD39 pKa = 4.08YY40 pKa = 10.35EE41 pKa = 4.09NSQLDD46 pKa = 3.44LSIPCRR52 pKa = 11.84TCIGCRR58 pKa = 11.84LDD60 pKa = 5.38QSRR63 pKa = 11.84QWADD67 pKa = 2.63RR68 pKa = 11.84MMIEE72 pKa = 5.16LDD74 pKa = 3.3HH75 pKa = 6.63SKK77 pKa = 10.95CGIFVTLTYY86 pKa = 10.75DD87 pKa = 3.19PAFAHH92 pKa = 6.76PCSIDD97 pKa = 3.27SDD99 pKa = 3.8DD100 pKa = 3.59RR101 pKa = 11.84VYY103 pKa = 11.56YY104 pKa = 10.75SLDD107 pKa = 3.31VRR109 pKa = 11.84DD110 pKa = 3.63TQLFNKK116 pKa = 9.26SLRR119 pKa = 11.84KK120 pKa = 9.82AFPDD124 pKa = 3.07RR125 pKa = 11.84KK126 pKa = 9.97IRR128 pKa = 11.84FFLAGEE134 pKa = 4.22YY135 pKa = 10.81GPSTWRR141 pKa = 11.84PHH143 pKa = 3.84YY144 pKa = 9.6HH145 pKa = 6.07AVYY148 pKa = 10.06FGLSIDD154 pKa = 4.72DD155 pKa = 5.26FPDD158 pKa = 3.76LMPLKK163 pKa = 9.2RR164 pKa = 11.84TSAGSLIYY172 pKa = 10.23TSEE175 pKa = 4.15KK176 pKa = 10.18LQKK179 pKa = 8.78LWKK182 pKa = 9.88RR183 pKa = 11.84GFISLSPVSWQTCAYY198 pKa = 8.04VARR201 pKa = 11.84YY202 pKa = 5.27TTKK205 pKa = 10.96KK206 pKa = 10.74LIDD209 pKa = 4.27SNPLMSDD216 pKa = 3.0YY217 pKa = 11.48DD218 pKa = 4.4LFNMHH223 pKa = 7.43PEE225 pKa = 3.91FMTCSRR231 pKa = 11.84KK232 pKa = 9.69PGLSGYY238 pKa = 10.31FYY240 pKa = 10.56DD241 pKa = 4.79DD242 pKa = 4.41HH243 pKa = 7.5PDD245 pKa = 3.63LVPDD249 pKa = 4.94SIDD252 pKa = 3.3DD253 pKa = 3.7FKK255 pKa = 11.36IYY257 pKa = 10.27IRR259 pKa = 11.84DD260 pKa = 3.68PDD262 pKa = 3.56GLKK265 pKa = 10.08PVKK268 pKa = 10.09SVKK271 pKa = 8.75TPKK274 pKa = 10.5YY275 pKa = 10.33LLKK278 pKa = 10.37QLQAVNPLLYY288 pKa = 10.51NEE290 pKa = 4.3YY291 pKa = 10.63VDD293 pKa = 3.7MQRR296 pKa = 11.84VSSEE300 pKa = 3.99DD301 pKa = 3.39SLLLKK306 pKa = 10.2LQRR309 pKa = 11.84TSLPEE314 pKa = 3.59AEE316 pKa = 4.78YY317 pKa = 10.51ILSEE321 pKa = 4.33EE322 pKa = 4.17FTKK325 pKa = 10.91QNSAKK330 pKa = 10.45KK331 pKa = 10.45LIRR334 pKa = 11.84SDD336 pKa = 3.5LL337 pKa = 3.76
Molecular weight: 39.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1493 |
82 |
570 |
248.8 |
27.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.167 ± 1.321 | 1.273 ± 0.45 |
7.77 ± 1.217 | 3.885 ± 0.381 |
5.894 ± 0.789 | 5.894 ± 1.034 |
1.741 ± 0.428 | 4.287 ± 0.362 |
3.952 ± 0.9 | 9.109 ± 0.987 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.626 | 5.023 ± 0.861 |
6.095 ± 0.422 | 3.215 ± 0.589 |
4.756 ± 0.627 | 10.181 ± 1.775 |
5.291 ± 0.92 | 5.76 ± 0.584 |
1.407 ± 0.395 | 5.023 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
