
Tremella mesenterica (strain ATCC 24925 / CBS 8224 / DSM 1558 / NBRC 9311 / NRRL Y-6157 / RJB 2259-6 / UBC 559-6) (Jelly fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Tremellaceae; Tremella; Tremella mesenterica
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
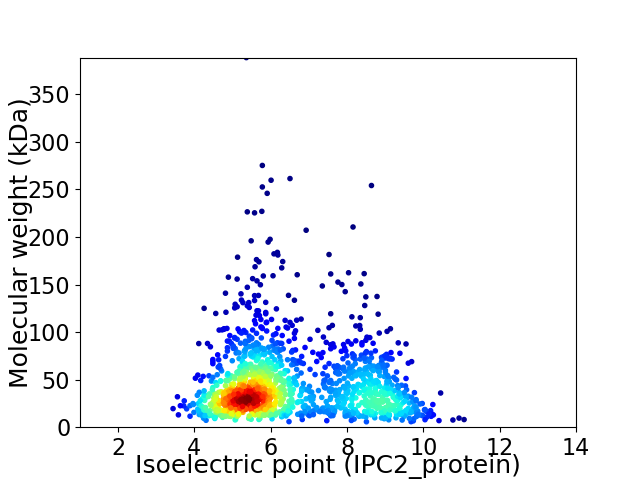
Virtual 2D-PAGE plot for 1453 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
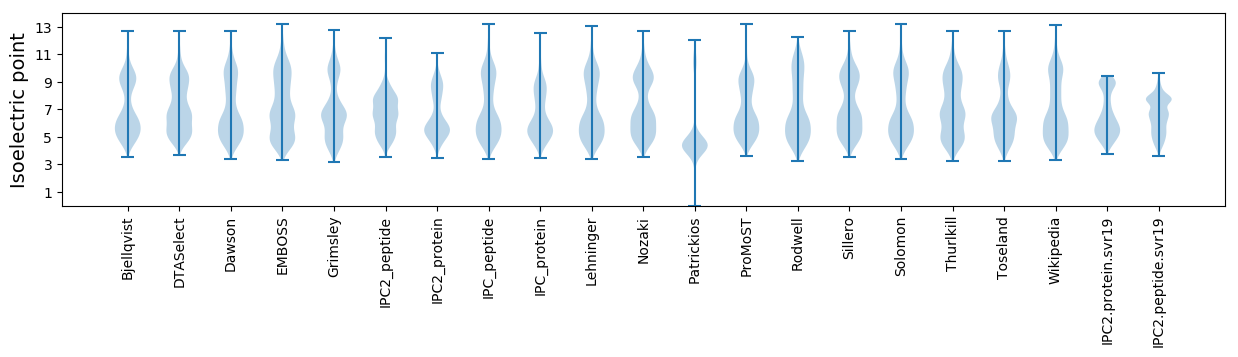
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7SD52|R7SD52_TREMS Pyridoxal 5'-phosphate synthase OS=Tremella mesenterica (strain ATCC 24925 / CBS 8224 / DSM 1558 / NBRC 9311 / NRRL Y-6157 / RJB 2259-6 / UBC 559-6) OX=578456 GN=TREMEDRAFT_70272 PE=3 SV=1
MM1 pKa = 7.85SSDD4 pKa = 3.56QPKK7 pKa = 8.09TTSQDD12 pKa = 3.47EE13 pKa = 4.3ILSFPFPTNADD24 pKa = 3.31LSLPGPSQFNTTLPSDD40 pKa = 4.09PFSMHH45 pKa = 6.87GLPPTISAEE54 pKa = 4.02FDD56 pKa = 3.8SNPIDD61 pKa = 3.44MDD63 pKa = 4.14SPSMSFLNNMNFGDD77 pKa = 4.63NPDD80 pKa = 3.63QGSGTGFNMNQYY92 pKa = 10.12EE93 pKa = 4.45MPSPNSFFPFGFSGGQSGTGNHH115 pKa = 5.83LQVGQGGVGNNFQVGQSGIPNSFGQNQNGYY145 pKa = 9.56PYY147 pKa = 10.03GQNNTGIPFGDD158 pKa = 3.43GTGFIPQTGRR168 pKa = 11.84TPGDD172 pKa = 3.37TYY174 pKa = 11.49GYY176 pKa = 9.51GQNLNWDD183 pKa = 3.92TPGNPIGRR191 pKa = 11.84GVDD194 pKa = 3.17VGAGFTSGTGGINEE208 pKa = 4.54GGGGSTGFTPP218 pKa = 4.95
MM1 pKa = 7.85SSDD4 pKa = 3.56QPKK7 pKa = 8.09TTSQDD12 pKa = 3.47EE13 pKa = 4.3ILSFPFPTNADD24 pKa = 3.31LSLPGPSQFNTTLPSDD40 pKa = 4.09PFSMHH45 pKa = 6.87GLPPTISAEE54 pKa = 4.02FDD56 pKa = 3.8SNPIDD61 pKa = 3.44MDD63 pKa = 4.14SPSMSFLNNMNFGDD77 pKa = 4.63NPDD80 pKa = 3.63QGSGTGFNMNQYY92 pKa = 10.12EE93 pKa = 4.45MPSPNSFFPFGFSGGQSGTGNHH115 pKa = 5.83LQVGQGGVGNNFQVGQSGIPNSFGQNQNGYY145 pKa = 9.56PYY147 pKa = 10.03GQNNTGIPFGDD158 pKa = 3.43GTGFIPQTGRR168 pKa = 11.84TPGDD172 pKa = 3.37TYY174 pKa = 11.49GYY176 pKa = 9.51GQNLNWDD183 pKa = 3.92TPGNPIGRR191 pKa = 11.84GVDD194 pKa = 3.17VGAGFTSGTGGINEE208 pKa = 4.54GGGGSTGFTPP218 pKa = 4.95
Molecular weight: 22.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7SC37|R7SC37_TREMS MFS domain-containing protein OS=Tremella mesenterica (strain ATCC 24925 / CBS 8224 / DSM 1558 / NBRC 9311 / NRRL Y-6157 / RJB 2259-6 / UBC 559-6) OX=578456 GN=TREMEDRAFT_34999 PE=4 SV=1
PP1 pKa = 6.32TQSSLAKK8 pKa = 9.93VRR10 pKa = 11.84EE11 pKa = 4.24KK12 pKa = 10.51FVPPPTRR19 pKa = 11.84PSPAMSRR26 pKa = 11.84PATAMSRR33 pKa = 11.84PPTAMSRR40 pKa = 11.84PATAMSRR47 pKa = 11.84PPTAMSRR54 pKa = 11.84PATAMSRR61 pKa = 11.84PPTAMSRR68 pKa = 11.84SSTALSRR75 pKa = 11.84PPTAMSRR82 pKa = 11.84PATAMSRR89 pKa = 11.84PAA91 pKa = 3.71
PP1 pKa = 6.32TQSSLAKK8 pKa = 9.93VRR10 pKa = 11.84EE11 pKa = 4.24KK12 pKa = 10.51FVPPPTRR19 pKa = 11.84PSPAMSRR26 pKa = 11.84PATAMSRR33 pKa = 11.84PPTAMSRR40 pKa = 11.84PATAMSRR47 pKa = 11.84PPTAMSRR54 pKa = 11.84PATAMSRR61 pKa = 11.84PPTAMSRR68 pKa = 11.84SSTALSRR75 pKa = 11.84PPTAMSRR82 pKa = 11.84PATAMSRR89 pKa = 11.84PAA91 pKa = 3.71
Molecular weight: 9.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
621631 |
51 |
3440 |
427.8 |
47.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.767 ± 0.076 | 1.197 ± 0.029 |
5.608 ± 0.051 | 6.582 ± 0.068 |
3.21 ± 0.034 | 6.905 ± 0.063 |
2.51 ± 0.038 | 4.788 ± 0.047 |
4.831 ± 0.058 | 8.982 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.025 | 3.518 ± 0.035 |
6.916 ± 0.093 | 3.873 ± 0.044 |
6.506 ± 0.059 | 9.366 ± 0.087 |
6.338 ± 0.051 | 6.097 ± 0.044 |
1.447 ± 0.023 | 2.37 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
