
Taro vein chlorosis virus (TAVCV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
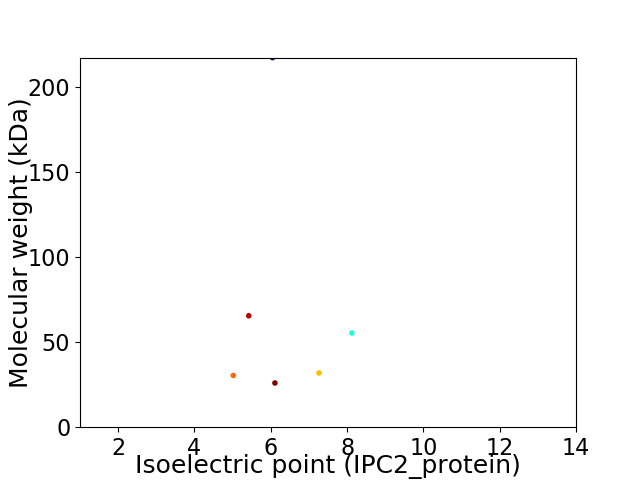
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5GA90|NCAP_TAVCV Nucleoprotein OS=Taro vein chlorosis virus OX=303300 GN=N PE=3 SV=1
MM1 pKa = 7.64INTSRR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84NKK12 pKa = 9.24PATPAPEE19 pKa = 4.68LIKK22 pKa = 10.91AEE24 pKa = 4.2EE25 pKa = 4.21NPSDD29 pKa = 3.3IHH31 pKa = 7.52EE32 pKa = 5.15DD33 pKa = 3.54FADD36 pKa = 4.01LPEE39 pKa = 4.8PLLLSEE45 pKa = 4.9ARR47 pKa = 11.84AEE49 pKa = 4.0NMPAVGSAISDD60 pKa = 3.79LLKK63 pKa = 10.69VEE65 pKa = 4.62RR66 pKa = 11.84AAPLLIGNTVEE77 pKa = 4.01EE78 pKa = 4.78MVTGRR83 pKa = 11.84LADD86 pKa = 3.95RR87 pKa = 11.84GLGEE91 pKa = 3.77LTEE94 pKa = 4.11RR95 pKa = 11.84EE96 pKa = 4.31KK97 pKa = 10.76IILAIGIHH105 pKa = 6.29CGEE108 pKa = 4.46SSMEE112 pKa = 3.89HH113 pKa = 5.66MNFLVTKK120 pKa = 9.84RR121 pKa = 11.84WITEE125 pKa = 3.81EE126 pKa = 5.13LKK128 pKa = 9.25TQLTSLAATTRR139 pKa = 11.84ALTEE143 pKa = 4.23AGSLHH148 pKa = 5.82RR149 pKa = 11.84TYY151 pKa = 11.64ALLQSPDD158 pKa = 3.27QAKK161 pKa = 10.01KK162 pKa = 10.58QEE164 pKa = 4.36ALDD167 pKa = 4.02AMVTPQVGIDD177 pKa = 3.13ITTLNHH183 pKa = 6.71EE184 pKa = 4.35GLEE187 pKa = 4.93DD188 pKa = 2.9IWQSYY193 pKa = 7.61SHH195 pKa = 7.17DD196 pKa = 3.82AKK198 pKa = 11.13VDD200 pKa = 3.44AVDD203 pKa = 4.6DD204 pKa = 3.88YY205 pKa = 11.77LRR207 pKa = 11.84NILNVDD213 pKa = 3.72PTPLYY218 pKa = 11.03AEE220 pKa = 5.0DD221 pKa = 2.99EE222 pKa = 4.47WGRR225 pKa = 11.84HH226 pKa = 4.39IAFIPRR232 pKa = 11.84WQLVAYY238 pKa = 8.3GKK240 pKa = 10.39NSDD243 pKa = 3.61QFKK246 pKa = 9.56TVYY249 pKa = 10.52ADD251 pKa = 3.68EE252 pKa = 4.68INYY255 pKa = 8.96QRR257 pKa = 11.84EE258 pKa = 4.0SLEE261 pKa = 4.26RR262 pKa = 11.84ILAKK266 pKa = 10.01RR267 pKa = 11.84VKK269 pKa = 10.65LNN271 pKa = 3.24
MM1 pKa = 7.64INTSRR6 pKa = 11.84RR7 pKa = 11.84SSRR10 pKa = 11.84NKK12 pKa = 9.24PATPAPEE19 pKa = 4.68LIKK22 pKa = 10.91AEE24 pKa = 4.2EE25 pKa = 4.21NPSDD29 pKa = 3.3IHH31 pKa = 7.52EE32 pKa = 5.15DD33 pKa = 3.54FADD36 pKa = 4.01LPEE39 pKa = 4.8PLLLSEE45 pKa = 4.9ARR47 pKa = 11.84AEE49 pKa = 4.0NMPAVGSAISDD60 pKa = 3.79LLKK63 pKa = 10.69VEE65 pKa = 4.62RR66 pKa = 11.84AAPLLIGNTVEE77 pKa = 4.01EE78 pKa = 4.78MVTGRR83 pKa = 11.84LADD86 pKa = 3.95RR87 pKa = 11.84GLGEE91 pKa = 3.77LTEE94 pKa = 4.11RR95 pKa = 11.84EE96 pKa = 4.31KK97 pKa = 10.76IILAIGIHH105 pKa = 6.29CGEE108 pKa = 4.46SSMEE112 pKa = 3.89HH113 pKa = 5.66MNFLVTKK120 pKa = 9.84RR121 pKa = 11.84WITEE125 pKa = 3.81EE126 pKa = 5.13LKK128 pKa = 9.25TQLTSLAATTRR139 pKa = 11.84ALTEE143 pKa = 4.23AGSLHH148 pKa = 5.82RR149 pKa = 11.84TYY151 pKa = 11.64ALLQSPDD158 pKa = 3.27QAKK161 pKa = 10.01KK162 pKa = 10.58QEE164 pKa = 4.36ALDD167 pKa = 4.02AMVTPQVGIDD177 pKa = 3.13ITTLNHH183 pKa = 6.71EE184 pKa = 4.35GLEE187 pKa = 4.93DD188 pKa = 2.9IWQSYY193 pKa = 7.61SHH195 pKa = 7.17DD196 pKa = 3.82AKK198 pKa = 11.13VDD200 pKa = 3.44AVDD203 pKa = 4.6DD204 pKa = 3.88YY205 pKa = 11.77LRR207 pKa = 11.84NILNVDD213 pKa = 3.72PTPLYY218 pKa = 11.03AEE220 pKa = 5.0DD221 pKa = 2.99EE222 pKa = 4.47WGRR225 pKa = 11.84HH226 pKa = 4.39IAFIPRR232 pKa = 11.84WQLVAYY238 pKa = 8.3GKK240 pKa = 10.39NSDD243 pKa = 3.61QFKK246 pKa = 9.56TVYY249 pKa = 10.52ADD251 pKa = 3.68EE252 pKa = 4.68INYY255 pKa = 8.96QRR257 pKa = 11.84EE258 pKa = 4.0SLEE261 pKa = 4.26RR262 pKa = 11.84ILAKK266 pKa = 10.01RR267 pKa = 11.84VKK269 pKa = 10.65LNN271 pKa = 3.24
Molecular weight: 30.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q5GA90|NCAP_TAVCV Nucleoprotein OS=Taro vein chlorosis virus OX=303300 GN=N PE=3 SV=1
MM1 pKa = 7.76SYY3 pKa = 10.99INIPDD8 pKa = 3.88DD9 pKa = 3.9VVSKK13 pKa = 11.28YY14 pKa = 11.03SDD16 pKa = 3.56DD17 pKa = 4.76LKK19 pKa = 10.48TFTQKK24 pKa = 10.81AGEE27 pKa = 4.19IPSSKK32 pKa = 10.85SLIPQTAYY40 pKa = 9.85TIAALKK46 pKa = 9.59TKK48 pKa = 10.58LKK50 pKa = 9.96FWEE53 pKa = 4.52VTAKK57 pKa = 10.45DD58 pKa = 4.23DD59 pKa = 3.84PTIASDD65 pKa = 3.21WAGVCTAITAGTFSATNLKK84 pKa = 8.11TVCEE88 pKa = 3.89LAFNLRR94 pKa = 11.84KK95 pKa = 9.12PHH97 pKa = 5.1EE98 pKa = 4.45TGNVFIHH105 pKa = 6.13TVPSDD110 pKa = 3.24WTSSISTDD118 pKa = 3.75SVDD121 pKa = 3.62TTPIPATEE129 pKa = 4.03SDD131 pKa = 3.46ATLSTVSAAVQSGAAEE147 pKa = 4.15DD148 pKa = 3.87AATKK152 pKa = 10.57AKK154 pKa = 10.4AISFLCCALIRR165 pKa = 11.84LSVKK169 pKa = 10.05EE170 pKa = 4.33PSHH173 pKa = 7.8IMTAITSIRR182 pKa = 11.84QRR184 pKa = 11.84FGSLYY189 pKa = 9.97GLASATLNAITFTRR203 pKa = 11.84QQLSRR208 pKa = 11.84IKK210 pKa = 10.53QGIEE214 pKa = 3.87TYY216 pKa = 10.58SLARR220 pKa = 11.84GTIFYY225 pKa = 9.36YY226 pKa = 10.61VRR228 pKa = 11.84YY229 pKa = 10.1ADD231 pKa = 3.66TTYY234 pKa = 11.11GSSDD238 pKa = 2.83KK239 pKa = 11.18SYY241 pKa = 10.43GVCRR245 pKa = 11.84FLLFQHH251 pKa = 6.78LEE253 pKa = 4.08LEE255 pKa = 4.52GMHH258 pKa = 6.72IYY260 pKa = 11.13KK261 pKa = 9.84MILALLTEE269 pKa = 4.88WSTVPIGLLLTWIRR283 pKa = 11.84NPKK286 pKa = 9.13SALAVVEE293 pKa = 4.21IKK295 pKa = 10.93NIITNFDD302 pKa = 3.15KK303 pKa = 11.43AGVDD307 pKa = 3.64KK308 pKa = 10.51SWKK311 pKa = 7.56YY312 pKa = 11.61SRR314 pKa = 11.84MIDD317 pKa = 3.03NTFFLNISSRR327 pKa = 11.84RR328 pKa = 11.84NVYY331 pKa = 8.77MCALLASLNKK341 pKa = 9.57RR342 pKa = 11.84HH343 pKa = 5.69VPQGVGDD350 pKa = 3.81YY351 pKa = 11.11ADD353 pKa = 3.9PRR355 pKa = 11.84NIAVIKK361 pKa = 11.03AMDD364 pKa = 4.01AAVKK368 pKa = 9.95NQVAIDD374 pKa = 3.61VTLVEE379 pKa = 5.09RR380 pKa = 11.84IYY382 pKa = 11.04EE383 pKa = 4.18KK384 pKa = 11.28YY385 pKa = 10.49LISAGSTDD393 pKa = 3.7AGTAYY398 pKa = 9.7TLSRR402 pKa = 11.84GTKK405 pKa = 8.84RR406 pKa = 11.84PNPAVFMSHH415 pKa = 5.34QQAEE419 pKa = 4.19GHH421 pKa = 5.16PTKK424 pKa = 10.69KK425 pKa = 9.45RR426 pKa = 11.84TWKK429 pKa = 10.86SMVPPPPTLQLEE441 pKa = 4.43DD442 pKa = 3.43QQQRR446 pKa = 11.84RR447 pKa = 11.84LSPEE451 pKa = 3.47LCEE454 pKa = 4.95DD455 pKa = 3.9CFSEE459 pKa = 4.43LLRR462 pKa = 11.84GKK464 pKa = 8.42HH465 pKa = 5.69HH466 pKa = 7.54RR467 pKa = 11.84ILPPPVIPSSSSEE480 pKa = 3.87VLFSKK485 pKa = 10.47SRR487 pKa = 11.84IMEE490 pKa = 3.81ISGGCSFDD498 pKa = 4.28PIGFF502 pKa = 3.93
MM1 pKa = 7.76SYY3 pKa = 10.99INIPDD8 pKa = 3.88DD9 pKa = 3.9VVSKK13 pKa = 11.28YY14 pKa = 11.03SDD16 pKa = 3.56DD17 pKa = 4.76LKK19 pKa = 10.48TFTQKK24 pKa = 10.81AGEE27 pKa = 4.19IPSSKK32 pKa = 10.85SLIPQTAYY40 pKa = 9.85TIAALKK46 pKa = 9.59TKK48 pKa = 10.58LKK50 pKa = 9.96FWEE53 pKa = 4.52VTAKK57 pKa = 10.45DD58 pKa = 4.23DD59 pKa = 3.84PTIASDD65 pKa = 3.21WAGVCTAITAGTFSATNLKK84 pKa = 8.11TVCEE88 pKa = 3.89LAFNLRR94 pKa = 11.84KK95 pKa = 9.12PHH97 pKa = 5.1EE98 pKa = 4.45TGNVFIHH105 pKa = 6.13TVPSDD110 pKa = 3.24WTSSISTDD118 pKa = 3.75SVDD121 pKa = 3.62TTPIPATEE129 pKa = 4.03SDD131 pKa = 3.46ATLSTVSAAVQSGAAEE147 pKa = 4.15DD148 pKa = 3.87AATKK152 pKa = 10.57AKK154 pKa = 10.4AISFLCCALIRR165 pKa = 11.84LSVKK169 pKa = 10.05EE170 pKa = 4.33PSHH173 pKa = 7.8IMTAITSIRR182 pKa = 11.84QRR184 pKa = 11.84FGSLYY189 pKa = 9.97GLASATLNAITFTRR203 pKa = 11.84QQLSRR208 pKa = 11.84IKK210 pKa = 10.53QGIEE214 pKa = 3.87TYY216 pKa = 10.58SLARR220 pKa = 11.84GTIFYY225 pKa = 9.36YY226 pKa = 10.61VRR228 pKa = 11.84YY229 pKa = 10.1ADD231 pKa = 3.66TTYY234 pKa = 11.11GSSDD238 pKa = 2.83KK239 pKa = 11.18SYY241 pKa = 10.43GVCRR245 pKa = 11.84FLLFQHH251 pKa = 6.78LEE253 pKa = 4.08LEE255 pKa = 4.52GMHH258 pKa = 6.72IYY260 pKa = 11.13KK261 pKa = 9.84MILALLTEE269 pKa = 4.88WSTVPIGLLLTWIRR283 pKa = 11.84NPKK286 pKa = 9.13SALAVVEE293 pKa = 4.21IKK295 pKa = 10.93NIITNFDD302 pKa = 3.15KK303 pKa = 11.43AGVDD307 pKa = 3.64KK308 pKa = 10.51SWKK311 pKa = 7.56YY312 pKa = 11.61SRR314 pKa = 11.84MIDD317 pKa = 3.03NTFFLNISSRR327 pKa = 11.84RR328 pKa = 11.84NVYY331 pKa = 8.77MCALLASLNKK341 pKa = 9.57RR342 pKa = 11.84HH343 pKa = 5.69VPQGVGDD350 pKa = 3.81YY351 pKa = 11.11ADD353 pKa = 3.9PRR355 pKa = 11.84NIAVIKK361 pKa = 11.03AMDD364 pKa = 4.01AAVKK368 pKa = 9.95NQVAIDD374 pKa = 3.61VTLVEE379 pKa = 5.09RR380 pKa = 11.84IYY382 pKa = 11.04EE383 pKa = 4.18KK384 pKa = 11.28YY385 pKa = 10.49LISAGSTDD393 pKa = 3.7AGTAYY398 pKa = 9.7TLSRR402 pKa = 11.84GTKK405 pKa = 8.84RR406 pKa = 11.84PNPAVFMSHH415 pKa = 5.34QQAEE419 pKa = 4.19GHH421 pKa = 5.16PTKK424 pKa = 10.69KK425 pKa = 9.45RR426 pKa = 11.84TWKK429 pKa = 10.86SMVPPPPTLQLEE441 pKa = 4.43DD442 pKa = 3.43QQQRR446 pKa = 11.84RR447 pKa = 11.84LSPEE451 pKa = 3.47LCEE454 pKa = 4.95DD455 pKa = 3.9CFSEE459 pKa = 4.43LLRR462 pKa = 11.84GKK464 pKa = 8.42HH465 pKa = 5.69HH466 pKa = 7.54RR467 pKa = 11.84ILPPPVIPSSSSEE480 pKa = 3.87VLFSKK485 pKa = 10.47SRR487 pKa = 11.84IMEE490 pKa = 3.81ISGGCSFDD498 pKa = 4.28PIGFF502 pKa = 3.93
Molecular weight: 55.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3811 |
235 |
1928 |
635.2 |
71.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.589 ± 0.878 | 1.811 ± 0.183 |
5.668 ± 0.514 | 5.432 ± 0.492 |
2.939 ± 0.238 | 6.166 ± 0.452 |
2.86 ± 0.289 | 7.373 ± 0.392 |
5.379 ± 0.236 | 10.575 ± 0.788 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.372 | 3.7 ± 0.252 |
4.854 ± 0.328 | 3.044 ± 0.079 |
4.933 ± 0.526 | 8.948 ± 0.428 |
6.534 ± 0.796 | 6.429 ± 0.309 |
1.863 ± 0.295 | 3.332 ± 0.127 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
