
Meles meles circovirus-like virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins
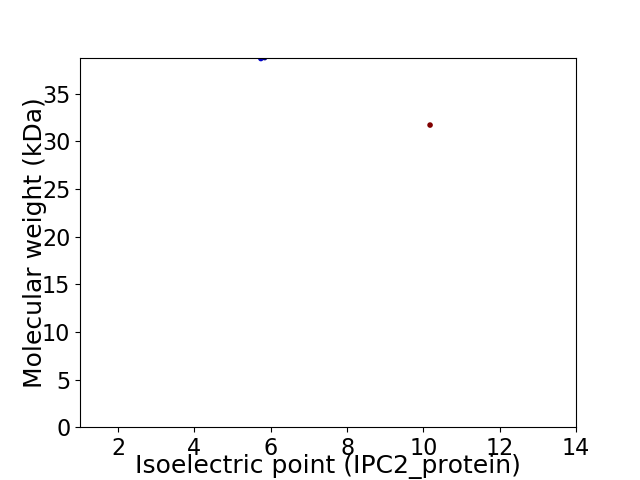
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9LQV7|G9LQV7_9CIRC Capsid protein OS=Meles meles circovirus-like virus OX=1128073 PE=4 SV=1
MM1 pKa = 7.24ATRR4 pKa = 11.84PEE6 pKa = 3.99ALHH9 pKa = 6.26WCFTLNNPTIDD20 pKa = 3.53EE21 pKa = 4.03QQIIDD26 pKa = 3.84EE27 pKa = 4.48LEE29 pKa = 3.94EE30 pKa = 3.99FGIEE34 pKa = 3.99YY35 pKa = 10.58AVFQLEE41 pKa = 4.15EE42 pKa = 4.56GEE44 pKa = 4.35NNTPHH49 pKa = 5.72FQGYY53 pKa = 8.35VALTVKK59 pKa = 10.31KK60 pKa = 10.19RR61 pKa = 11.84LTAVRR66 pKa = 11.84TLFGGVAHH74 pKa = 6.82WEE76 pKa = 4.18ICRR79 pKa = 11.84GTPTQNRR86 pKa = 11.84DD87 pKa = 3.08YY88 pKa = 9.93CTKK91 pKa = 10.43ADD93 pKa = 3.59TRR95 pKa = 11.84IGDD98 pKa = 3.7FVEE101 pKa = 4.93IGTLPAPKK109 pKa = 10.04GARR112 pKa = 11.84QDD114 pKa = 3.96LVQLHH119 pKa = 6.25SALKK123 pKa = 10.71DD124 pKa = 3.31GLTQAQYY131 pKa = 11.02VDD133 pKa = 3.44QFFSIFVRR141 pKa = 11.84HH142 pKa = 6.35PNLVSNYY149 pKa = 7.54VAATISEE156 pKa = 4.33RR157 pKa = 11.84DD158 pKa = 3.27PANPVSTWLLIGTPGTGKK176 pKa = 10.26SRR178 pKa = 11.84LARR181 pKa = 11.84AIGKK185 pKa = 8.76NLGDD189 pKa = 3.56GAIFEE194 pKa = 4.64HH195 pKa = 7.27SLGKK199 pKa = 9.81WFDD202 pKa = 3.74GYY204 pKa = 11.03VGQRR208 pKa = 11.84TVLFDD213 pKa = 4.08DD214 pKa = 4.8FCGSSLPFSQFKK226 pKa = 10.17RR227 pKa = 11.84ICDD230 pKa = 3.78RR231 pKa = 11.84YY232 pKa = 8.76PVRR235 pKa = 11.84VEE237 pKa = 4.87LKK239 pKa = 10.74GLSCPMAATNFLITTNQDD257 pKa = 2.68PKK259 pKa = 10.37TWWKK263 pKa = 11.3DD264 pKa = 3.42EE265 pKa = 3.83VTGQHH270 pKa = 4.7GHH272 pKa = 5.92AAITRR277 pKa = 11.84RR278 pKa = 11.84IGRR281 pKa = 11.84VLFFFAEE288 pKa = 4.24NTFRR292 pKa = 11.84SYY294 pKa = 11.23PSYY297 pKa = 10.87NHH299 pKa = 6.47YY300 pKa = 9.95ATDD303 pKa = 3.84CLRR306 pKa = 11.84PRR308 pKa = 11.84IDD310 pKa = 3.42GEE312 pKa = 4.45TYY314 pKa = 10.11HH315 pKa = 6.35YY316 pKa = 9.82FSPAQTIVYY325 pKa = 8.47DD326 pKa = 4.01AQGEE330 pKa = 4.47AILPPEE336 pKa = 4.14ILRR339 pKa = 11.84AEE341 pKa = 4.38VQQ343 pKa = 3.11
MM1 pKa = 7.24ATRR4 pKa = 11.84PEE6 pKa = 3.99ALHH9 pKa = 6.26WCFTLNNPTIDD20 pKa = 3.53EE21 pKa = 4.03QQIIDD26 pKa = 3.84EE27 pKa = 4.48LEE29 pKa = 3.94EE30 pKa = 3.99FGIEE34 pKa = 3.99YY35 pKa = 10.58AVFQLEE41 pKa = 4.15EE42 pKa = 4.56GEE44 pKa = 4.35NNTPHH49 pKa = 5.72FQGYY53 pKa = 8.35VALTVKK59 pKa = 10.31KK60 pKa = 10.19RR61 pKa = 11.84LTAVRR66 pKa = 11.84TLFGGVAHH74 pKa = 6.82WEE76 pKa = 4.18ICRR79 pKa = 11.84GTPTQNRR86 pKa = 11.84DD87 pKa = 3.08YY88 pKa = 9.93CTKK91 pKa = 10.43ADD93 pKa = 3.59TRR95 pKa = 11.84IGDD98 pKa = 3.7FVEE101 pKa = 4.93IGTLPAPKK109 pKa = 10.04GARR112 pKa = 11.84QDD114 pKa = 3.96LVQLHH119 pKa = 6.25SALKK123 pKa = 10.71DD124 pKa = 3.31GLTQAQYY131 pKa = 11.02VDD133 pKa = 3.44QFFSIFVRR141 pKa = 11.84HH142 pKa = 6.35PNLVSNYY149 pKa = 7.54VAATISEE156 pKa = 4.33RR157 pKa = 11.84DD158 pKa = 3.27PANPVSTWLLIGTPGTGKK176 pKa = 10.26SRR178 pKa = 11.84LARR181 pKa = 11.84AIGKK185 pKa = 8.76NLGDD189 pKa = 3.56GAIFEE194 pKa = 4.64HH195 pKa = 7.27SLGKK199 pKa = 9.81WFDD202 pKa = 3.74GYY204 pKa = 11.03VGQRR208 pKa = 11.84TVLFDD213 pKa = 4.08DD214 pKa = 4.8FCGSSLPFSQFKK226 pKa = 10.17RR227 pKa = 11.84ICDD230 pKa = 3.78RR231 pKa = 11.84YY232 pKa = 8.76PVRR235 pKa = 11.84VEE237 pKa = 4.87LKK239 pKa = 10.74GLSCPMAATNFLITTNQDD257 pKa = 2.68PKK259 pKa = 10.37TWWKK263 pKa = 11.3DD264 pKa = 3.42EE265 pKa = 3.83VTGQHH270 pKa = 4.7GHH272 pKa = 5.92AAITRR277 pKa = 11.84RR278 pKa = 11.84IGRR281 pKa = 11.84VLFFFAEE288 pKa = 4.24NTFRR292 pKa = 11.84SYY294 pKa = 11.23PSYY297 pKa = 10.87NHH299 pKa = 6.47YY300 pKa = 9.95ATDD303 pKa = 3.84CLRR306 pKa = 11.84PRR308 pKa = 11.84IDD310 pKa = 3.42GEE312 pKa = 4.45TYY314 pKa = 10.11HH315 pKa = 6.35YY316 pKa = 9.82FSPAQTIVYY325 pKa = 8.47DD326 pKa = 4.01AQGEE330 pKa = 4.47AILPPEE336 pKa = 4.14ILRR339 pKa = 11.84AEE341 pKa = 4.38VQQ343 pKa = 3.11
Molecular weight: 38.71 kDa
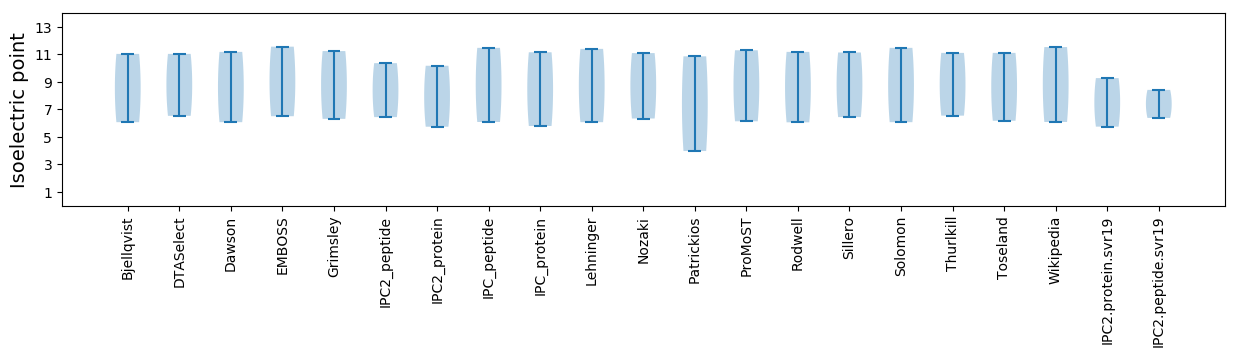
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9LQV7|G9LQV7_9CIRC Capsid protein OS=Meles meles circovirus-like virus OX=1128073 PE=4 SV=1
MM1 pKa = 7.47GRR3 pKa = 11.84RR4 pKa = 11.84ITTFRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SSTMRR18 pKa = 11.84KK19 pKa = 9.1ARR21 pKa = 11.84PSYY24 pKa = 10.06RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SSARR31 pKa = 11.84KK32 pKa = 7.87YY33 pKa = 9.04SKK35 pKa = 10.13KK36 pKa = 9.68RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84GMQRR44 pKa = 11.84IKK46 pKa = 9.78TSRR49 pKa = 11.84WPVRR53 pKa = 11.84NIGGDD58 pKa = 3.11RR59 pKa = 11.84SFCKK63 pKa = 10.12LRR65 pKa = 11.84YY66 pKa = 9.02VKK68 pKa = 10.66GSAFDD73 pKa = 3.58IPEE76 pKa = 4.34GAASLSQLQVMNVGAYY92 pKa = 9.27PGASNLLQSMTLNGVMGEE110 pKa = 4.3TPNLSTMGALYY121 pKa = 9.48TRR123 pKa = 11.84YY124 pKa = 9.87RR125 pKa = 11.84IRR127 pKa = 11.84GIKK130 pKa = 9.28IRR132 pKa = 11.84LTYY135 pKa = 8.28WQLGGAPVILFTNAAPDD152 pKa = 3.82VNNLTVASTGPNPAFITPNITTTTEE177 pKa = 3.89QRR179 pKa = 11.84WAKK182 pKa = 10.51YY183 pKa = 9.36RR184 pKa = 11.84VCQATANGGKK194 pKa = 8.19ATSVSAYY201 pKa = 9.28YY202 pKa = 10.34SVNRR206 pKa = 11.84VQGPDD211 pKa = 3.27TIVRR215 pKa = 11.84NDD217 pKa = 3.42RR218 pKa = 11.84DD219 pKa = 3.93YY220 pKa = 11.31TGEE223 pKa = 4.03MQPATPYY230 pKa = 9.53FANSSGVADD239 pKa = 4.39TPQFSPWMQWGIATLSGTPAVAGTQGVIKK268 pKa = 10.64VEE270 pKa = 3.8QTVYY274 pKa = 11.2CEE276 pKa = 4.08FFGKK280 pKa = 10.49RR281 pKa = 11.84IQTQQ285 pKa = 2.83
MM1 pKa = 7.47GRR3 pKa = 11.84RR4 pKa = 11.84ITTFRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SSTMRR18 pKa = 11.84KK19 pKa = 9.1ARR21 pKa = 11.84PSYY24 pKa = 10.06RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SSARR31 pKa = 11.84KK32 pKa = 7.87YY33 pKa = 9.04SKK35 pKa = 10.13KK36 pKa = 9.68RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84GMQRR44 pKa = 11.84IKK46 pKa = 9.78TSRR49 pKa = 11.84WPVRR53 pKa = 11.84NIGGDD58 pKa = 3.11RR59 pKa = 11.84SFCKK63 pKa = 10.12LRR65 pKa = 11.84YY66 pKa = 9.02VKK68 pKa = 10.66GSAFDD73 pKa = 3.58IPEE76 pKa = 4.34GAASLSQLQVMNVGAYY92 pKa = 9.27PGASNLLQSMTLNGVMGEE110 pKa = 4.3TPNLSTMGALYY121 pKa = 9.48TRR123 pKa = 11.84YY124 pKa = 9.87RR125 pKa = 11.84IRR127 pKa = 11.84GIKK130 pKa = 9.28IRR132 pKa = 11.84LTYY135 pKa = 8.28WQLGGAPVILFTNAAPDD152 pKa = 3.82VNNLTVASTGPNPAFITPNITTTTEE177 pKa = 3.89QRR179 pKa = 11.84WAKK182 pKa = 10.51YY183 pKa = 9.36RR184 pKa = 11.84VCQATANGGKK194 pKa = 8.19ATSVSAYY201 pKa = 9.28YY202 pKa = 10.34SVNRR206 pKa = 11.84VQGPDD211 pKa = 3.27TIVRR215 pKa = 11.84NDD217 pKa = 3.42RR218 pKa = 11.84DD219 pKa = 3.93YY220 pKa = 11.31TGEE223 pKa = 4.03MQPATPYY230 pKa = 9.53FANSSGVADD239 pKa = 4.39TPQFSPWMQWGIATLSGTPAVAGTQGVIKK268 pKa = 10.64VEE270 pKa = 3.8QTVYY274 pKa = 11.2CEE276 pKa = 4.08FFGKK280 pKa = 10.49RR281 pKa = 11.84IQTQQ285 pKa = 2.83
Molecular weight: 31.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
628 |
285 |
343 |
314.0 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.121 ± 0.182 | 1.592 ± 0.328 |
4.14 ± 1.023 | 4.14 ± 1.236 |
4.777 ± 0.983 | 8.121 ± 0.395 |
1.592 ± 0.967 | 5.414 ± 0.305 |
3.822 ± 0.236 | 6.369 ± 1.098 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.752 ± 0.854 | 4.299 ± 0.372 |
5.573 ± 0.025 | 5.096 ± 0.102 |
8.439 ± 1.48 | 5.892 ± 1.323 |
8.917 ± 0.551 | 6.051 ± 0.161 |
1.752 ± 0.002 | 4.14 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
