
Wenzhou tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.08
Get precalculated fractions of proteins
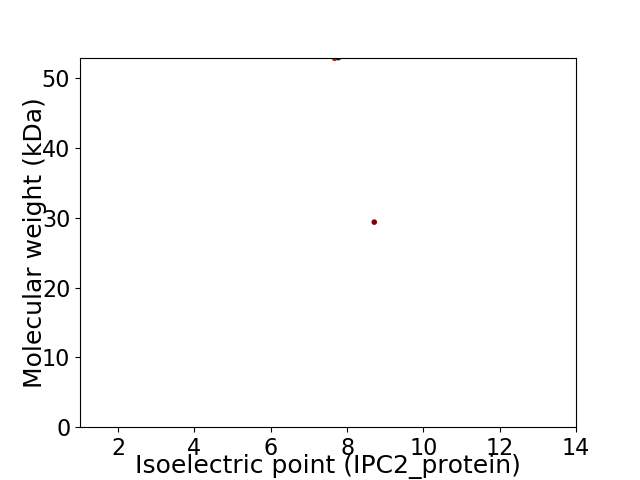
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH30|A0A1L3KH30_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 2 OX=1923672 PE=4 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84AITEE7 pKa = 3.59RR8 pKa = 11.84VLYY11 pKa = 9.93VRR13 pKa = 11.84RR14 pKa = 11.84DD15 pKa = 3.33GRR17 pKa = 11.84LTRR20 pKa = 11.84PPRR23 pKa = 11.84PEE25 pKa = 3.89PGFYY29 pKa = 10.59GQLSGIRR36 pKa = 11.84QRR38 pKa = 11.84LVRR41 pKa = 11.84KK42 pKa = 7.57ATPTPVVPLAEE53 pKa = 4.35YY54 pKa = 9.52PSLYY58 pKa = 9.99HH59 pKa = 5.52GRR61 pKa = 11.84KK62 pKa = 8.95RR63 pKa = 11.84KK64 pKa = 9.71IYY66 pKa = 8.43EE67 pKa = 3.83NAVNSLNEE75 pKa = 4.06RR76 pKa = 11.84GLTAKK81 pKa = 10.2DD82 pKa = 2.9AWVDD86 pKa = 3.55TFLKK90 pKa = 10.59AEE92 pKa = 4.27KK93 pKa = 10.88VNFDD97 pKa = 4.05SKK99 pKa = 11.44GDD101 pKa = 3.73PAPRR105 pKa = 11.84LISPRR110 pKa = 11.84GPRR113 pKa = 11.84YY114 pKa = 8.49NVEE117 pKa = 3.52VGRR120 pKa = 11.84YY121 pKa = 8.45LKK123 pKa = 10.82LFEE126 pKa = 5.2KK127 pKa = 10.61EE128 pKa = 3.64LFRR131 pKa = 11.84AFEE134 pKa = 4.0RR135 pKa = 11.84VFGYY139 pKa = 10.39KK140 pKa = 9.96VVVKK144 pKa = 10.1GLNAQQVGEE153 pKa = 4.84LLAQHH158 pKa = 6.31WGEE161 pKa = 3.75FRR163 pKa = 11.84EE164 pKa = 4.39PVAVGLDD171 pKa = 3.21ASRR174 pKa = 11.84FDD176 pKa = 3.42QHH178 pKa = 6.28VSRR181 pKa = 11.84DD182 pKa = 3.63ALEE185 pKa = 4.6WEE187 pKa = 4.14HH188 pKa = 7.38SVYY191 pKa = 10.89NAVFRR196 pKa = 11.84SEE198 pKa = 3.98RR199 pKa = 11.84LRR201 pKa = 11.84WLLRR205 pKa = 11.84MQLRR209 pKa = 11.84NRR211 pKa = 11.84CIARR215 pKa = 11.84VEE217 pKa = 4.08GRR219 pKa = 11.84RR220 pKa = 11.84FDD222 pKa = 3.62YY223 pKa = 8.44TTDD226 pKa = 3.39GVRR229 pKa = 11.84MSGDD233 pKa = 3.65LNTSLGNCLIMSSLVLRR250 pKa = 11.84YY251 pKa = 9.99LEE253 pKa = 4.02EE254 pKa = 5.4RR255 pKa = 11.84GVKK258 pKa = 9.9ARR260 pKa = 11.84LANNGDD266 pKa = 3.77DD267 pKa = 3.72CVVFCEE273 pKa = 4.82AADD276 pKa = 3.99LSKK279 pKa = 11.19LDD281 pKa = 5.28GLDD284 pKa = 3.13HH285 pKa = 6.48WFLRR289 pKa = 11.84AGFTLTRR296 pKa = 11.84EE297 pKa = 4.33DD298 pKa = 3.11PCYY301 pKa = 10.3EE302 pKa = 3.92LEE304 pKa = 4.4QVEE307 pKa = 5.11FCQFHH312 pKa = 6.34PVEE315 pKa = 4.82CANGWRR321 pKa = 11.84MVRR324 pKa = 11.84DD325 pKa = 3.42PRR327 pKa = 11.84VAMSKK332 pKa = 10.47DD333 pKa = 3.49CVSVVGWDD341 pKa = 3.34QEE343 pKa = 4.8SEE345 pKa = 4.55VKK347 pKa = 10.19VWAAAVGQCGLSLTAGVPVWEE368 pKa = 4.19SWYY371 pKa = 10.42KK372 pKa = 9.52RR373 pKa = 11.84LVRR376 pKa = 11.84VGVEE380 pKa = 3.63RR381 pKa = 11.84DD382 pKa = 3.18SGVGEE387 pKa = 4.07RR388 pKa = 11.84VKK390 pKa = 10.85EE391 pKa = 3.97CGMAYY396 pKa = 10.2AAAGVQACPISDD408 pKa = 3.4ACRR411 pKa = 11.84YY412 pKa = 9.3SFYY415 pKa = 10.45RR416 pKa = 11.84AFGILPDD423 pKa = 3.73LQEE426 pKa = 4.17ALEE429 pKa = 4.34RR430 pKa = 11.84EE431 pKa = 4.26YY432 pKa = 11.44SADD435 pKa = 3.49YY436 pKa = 10.02TLAPIDD442 pKa = 4.18PMMSSHH448 pKa = 7.14IKK450 pKa = 9.89TIDD453 pKa = 3.24QLNPLTLHH461 pKa = 6.39NGSS464 pKa = 3.9
MM1 pKa = 7.49RR2 pKa = 11.84RR3 pKa = 11.84AITEE7 pKa = 3.59RR8 pKa = 11.84VLYY11 pKa = 9.93VRR13 pKa = 11.84RR14 pKa = 11.84DD15 pKa = 3.33GRR17 pKa = 11.84LTRR20 pKa = 11.84PPRR23 pKa = 11.84PEE25 pKa = 3.89PGFYY29 pKa = 10.59GQLSGIRR36 pKa = 11.84QRR38 pKa = 11.84LVRR41 pKa = 11.84KK42 pKa = 7.57ATPTPVVPLAEE53 pKa = 4.35YY54 pKa = 9.52PSLYY58 pKa = 9.99HH59 pKa = 5.52GRR61 pKa = 11.84KK62 pKa = 8.95RR63 pKa = 11.84KK64 pKa = 9.71IYY66 pKa = 8.43EE67 pKa = 3.83NAVNSLNEE75 pKa = 4.06RR76 pKa = 11.84GLTAKK81 pKa = 10.2DD82 pKa = 2.9AWVDD86 pKa = 3.55TFLKK90 pKa = 10.59AEE92 pKa = 4.27KK93 pKa = 10.88VNFDD97 pKa = 4.05SKK99 pKa = 11.44GDD101 pKa = 3.73PAPRR105 pKa = 11.84LISPRR110 pKa = 11.84GPRR113 pKa = 11.84YY114 pKa = 8.49NVEE117 pKa = 3.52VGRR120 pKa = 11.84YY121 pKa = 8.45LKK123 pKa = 10.82LFEE126 pKa = 5.2KK127 pKa = 10.61EE128 pKa = 3.64LFRR131 pKa = 11.84AFEE134 pKa = 4.0RR135 pKa = 11.84VFGYY139 pKa = 10.39KK140 pKa = 9.96VVVKK144 pKa = 10.1GLNAQQVGEE153 pKa = 4.84LLAQHH158 pKa = 6.31WGEE161 pKa = 3.75FRR163 pKa = 11.84EE164 pKa = 4.39PVAVGLDD171 pKa = 3.21ASRR174 pKa = 11.84FDD176 pKa = 3.42QHH178 pKa = 6.28VSRR181 pKa = 11.84DD182 pKa = 3.63ALEE185 pKa = 4.6WEE187 pKa = 4.14HH188 pKa = 7.38SVYY191 pKa = 10.89NAVFRR196 pKa = 11.84SEE198 pKa = 3.98RR199 pKa = 11.84LRR201 pKa = 11.84WLLRR205 pKa = 11.84MQLRR209 pKa = 11.84NRR211 pKa = 11.84CIARR215 pKa = 11.84VEE217 pKa = 4.08GRR219 pKa = 11.84RR220 pKa = 11.84FDD222 pKa = 3.62YY223 pKa = 8.44TTDD226 pKa = 3.39GVRR229 pKa = 11.84MSGDD233 pKa = 3.65LNTSLGNCLIMSSLVLRR250 pKa = 11.84YY251 pKa = 9.99LEE253 pKa = 4.02EE254 pKa = 5.4RR255 pKa = 11.84GVKK258 pKa = 9.9ARR260 pKa = 11.84LANNGDD266 pKa = 3.77DD267 pKa = 3.72CVVFCEE273 pKa = 4.82AADD276 pKa = 3.99LSKK279 pKa = 11.19LDD281 pKa = 5.28GLDD284 pKa = 3.13HH285 pKa = 6.48WFLRR289 pKa = 11.84AGFTLTRR296 pKa = 11.84EE297 pKa = 4.33DD298 pKa = 3.11PCYY301 pKa = 10.3EE302 pKa = 3.92LEE304 pKa = 4.4QVEE307 pKa = 5.11FCQFHH312 pKa = 6.34PVEE315 pKa = 4.82CANGWRR321 pKa = 11.84MVRR324 pKa = 11.84DD325 pKa = 3.42PRR327 pKa = 11.84VAMSKK332 pKa = 10.47DD333 pKa = 3.49CVSVVGWDD341 pKa = 3.34QEE343 pKa = 4.8SEE345 pKa = 4.55VKK347 pKa = 10.19VWAAAVGQCGLSLTAGVPVWEE368 pKa = 4.19SWYY371 pKa = 10.42KK372 pKa = 9.52RR373 pKa = 11.84LVRR376 pKa = 11.84VGVEE380 pKa = 3.63RR381 pKa = 11.84DD382 pKa = 3.18SGVGEE387 pKa = 4.07RR388 pKa = 11.84VKK390 pKa = 10.85EE391 pKa = 3.97CGMAYY396 pKa = 10.2AAAGVQACPISDD408 pKa = 3.4ACRR411 pKa = 11.84YY412 pKa = 9.3SFYY415 pKa = 10.45RR416 pKa = 11.84AFGILPDD423 pKa = 3.73LQEE426 pKa = 4.17ALEE429 pKa = 4.34RR430 pKa = 11.84EE431 pKa = 4.26YY432 pKa = 11.44SADD435 pKa = 3.49YY436 pKa = 10.02TLAPIDD442 pKa = 4.18PMMSSHH448 pKa = 7.14IKK450 pKa = 9.89TIDD453 pKa = 3.24QLNPLTLHH461 pKa = 6.39NGSS464 pKa = 3.9
Molecular weight: 52.86 kDa
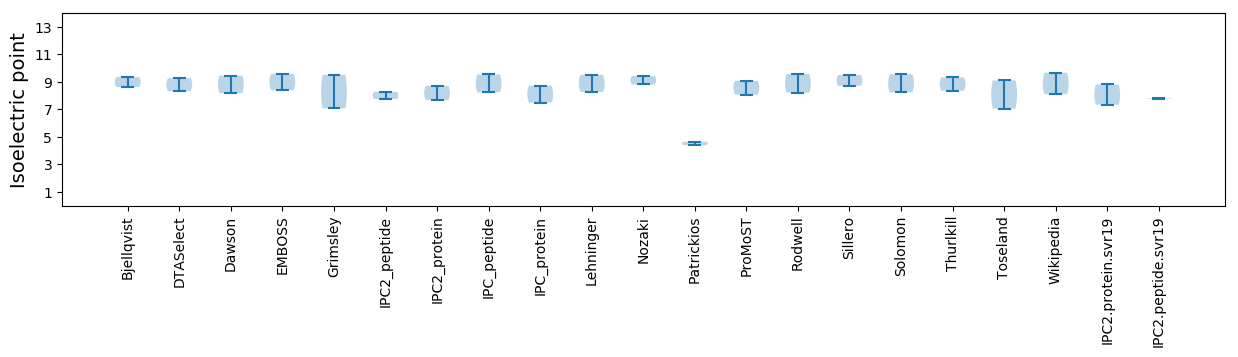
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH30|A0A1L3KH30_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 2 OX=1923672 PE=4 SV=1
MM1 pKa = 7.12ARR3 pKa = 11.84KK4 pKa = 9.04RR5 pKa = 11.84AKK7 pKa = 8.87QAQMPVSGGTRR18 pKa = 11.84RR19 pKa = 11.84NKK21 pKa = 10.61GYY23 pKa = 8.69TPTIKK28 pKa = 10.04GTGDD32 pKa = 3.3STIIGYY38 pKa = 9.41NGQLQPLIVNTSGRR52 pKa = 11.84GLNFRR57 pKa = 11.84QYY59 pKa = 10.69IGGFQEE65 pKa = 4.69AFAPGAIGPTIVARR79 pKa = 11.84YY80 pKa = 8.63ASCKK84 pKa = 9.63FLPGTTLRR92 pKa = 11.84WEE94 pKa = 4.23PSVSFSTSGRR104 pKa = 11.84VYY106 pKa = 11.03VGFTDD111 pKa = 3.62NPEE114 pKa = 3.61VTAYY118 pKa = 9.2MINLAVLTAADD129 pKa = 3.88PTNTGKK135 pKa = 9.81WNNYY139 pKa = 6.82EE140 pKa = 4.09SAVRR144 pKa = 11.84ALGSCISFPVWQEE157 pKa = 3.13TDD159 pKa = 2.83IPFPTKK165 pKa = 10.31LRR167 pKa = 11.84RR168 pKa = 11.84KK169 pKa = 10.07RR170 pKa = 11.84FDD172 pKa = 3.4TNQVFPNIAPAIDD185 pKa = 3.55TLDD188 pKa = 3.7RR189 pKa = 11.84CCQTTMFAAVVGVNTAVALTVGNFHH214 pKa = 6.39VHH216 pKa = 6.96DD217 pKa = 4.34KK218 pKa = 11.16VDD220 pKa = 3.6VEE222 pKa = 4.86GITPFITDD230 pKa = 3.11IDD232 pKa = 4.07TSLVASATPGSSSVQNLGHH251 pKa = 6.27SGGDD255 pKa = 3.11AVVGLGRR262 pKa = 11.84PHH264 pKa = 7.12PSAIAGYY271 pKa = 9.55NRR273 pKa = 11.84PLL275 pKa = 4.0
MM1 pKa = 7.12ARR3 pKa = 11.84KK4 pKa = 9.04RR5 pKa = 11.84AKK7 pKa = 8.87QAQMPVSGGTRR18 pKa = 11.84RR19 pKa = 11.84NKK21 pKa = 10.61GYY23 pKa = 8.69TPTIKK28 pKa = 10.04GTGDD32 pKa = 3.3STIIGYY38 pKa = 9.41NGQLQPLIVNTSGRR52 pKa = 11.84GLNFRR57 pKa = 11.84QYY59 pKa = 10.69IGGFQEE65 pKa = 4.69AFAPGAIGPTIVARR79 pKa = 11.84YY80 pKa = 8.63ASCKK84 pKa = 9.63FLPGTTLRR92 pKa = 11.84WEE94 pKa = 4.23PSVSFSTSGRR104 pKa = 11.84VYY106 pKa = 11.03VGFTDD111 pKa = 3.62NPEE114 pKa = 3.61VTAYY118 pKa = 9.2MINLAVLTAADD129 pKa = 3.88PTNTGKK135 pKa = 9.81WNNYY139 pKa = 6.82EE140 pKa = 4.09SAVRR144 pKa = 11.84ALGSCISFPVWQEE157 pKa = 3.13TDD159 pKa = 2.83IPFPTKK165 pKa = 10.31LRR167 pKa = 11.84RR168 pKa = 11.84KK169 pKa = 10.07RR170 pKa = 11.84FDD172 pKa = 3.4TNQVFPNIAPAIDD185 pKa = 3.55TLDD188 pKa = 3.7RR189 pKa = 11.84CCQTTMFAAVVGVNTAVALTVGNFHH214 pKa = 6.39VHH216 pKa = 6.96DD217 pKa = 4.34KK218 pKa = 11.16VDD220 pKa = 3.6VEE222 pKa = 4.86GITPFITDD230 pKa = 3.11IDD232 pKa = 4.07TSLVASATPGSSSVQNLGHH251 pKa = 6.27SGGDD255 pKa = 3.11AVVGLGRR262 pKa = 11.84PHH264 pKa = 7.12PSAIAGYY271 pKa = 9.55NRR273 pKa = 11.84PLL275 pKa = 4.0
Molecular weight: 29.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
739 |
275 |
464 |
369.5 |
41.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.39 ± 0.408 | 2.165 ± 0.413 |
5.142 ± 0.453 | 5.413 ± 1.878 |
4.195 ± 0.31 | 8.39 ± 1.042 |
1.624 ± 0.098 | 3.654 ± 1.258 |
3.654 ± 0.221 | 8.119 ± 1.549 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.759 ± 0.177 | 4.06 ± 0.811 |
5.548 ± 0.791 | 3.248 ± 0.226 |
8.66 ± 1.652 | 5.954 ± 0.344 |
5.683 ± 2.404 | 9.066 ± 0.409 |
1.759 ± 0.388 | 3.518 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
