
Porphyromonas asaccharolytica (strain ATCC 25260 / DSM 20707 / BCRC 10618 / JCM 6326 / LMG 13178 / VPI 4198) (Bacteroides asaccharolyticus)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Porphyromonadaceae; Porphyromonas; Porphyromonas asaccharolytica
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
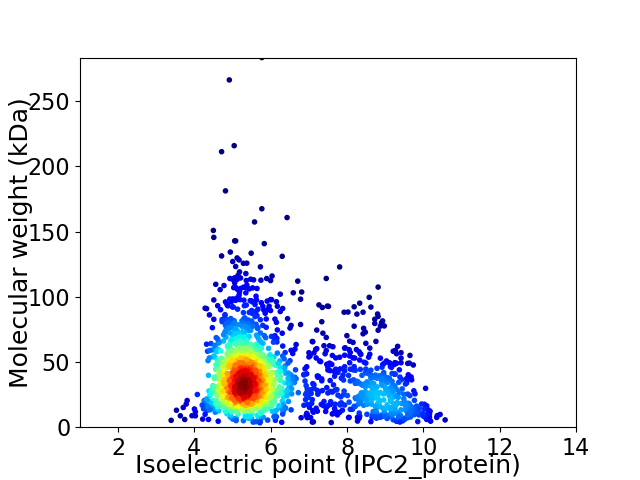
Virtual 2D-PAGE plot for 1672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F4KKC6|F4KKC6_PORAD Uncharacterized protein OS=Porphyromonas asaccharolytica (strain ATCC 25260 / DSM 20707 / BCRC 10618 / JCM 6326 / LMG 13178 / VPI 4198) OX=879243 GN=Poras_0908 PE=4 SV=1
MM1 pKa = 7.72KK2 pKa = 9.62NTGLYY7 pKa = 9.72FGSSTGHH14 pKa = 5.65VEE16 pKa = 4.83GIADD20 pKa = 4.58KK21 pKa = 10.81VAQLLGIDD29 pKa = 3.73AANVHH34 pKa = 5.87NVADD38 pKa = 4.16ATPEE42 pKa = 3.88SALAYY47 pKa = 10.07DD48 pKa = 4.23LLIFGSSTWGYY59 pKa = 11.66GDD61 pKa = 5.61LQDD64 pKa = 4.25DD65 pKa = 4.12WEE67 pKa = 4.79GFLPKK72 pKa = 10.14LVKK75 pKa = 9.92QDD77 pKa = 4.06LSGKK81 pKa = 8.56QFAVFGCGDD90 pKa = 3.45SSSYY94 pKa = 11.05PDD96 pKa = 3.71TFCDD100 pKa = 3.4AMAEE104 pKa = 4.0IAEE107 pKa = 4.33QVTAAGATLVGQIPAEE123 pKa = 4.61GYY125 pKa = 9.35SYY127 pKa = 10.86EE128 pKa = 4.05ATRR131 pKa = 11.84CEE133 pKa = 4.32VDD135 pKa = 3.35GQLIGCCLDD144 pKa = 3.73EE145 pKa = 6.29DD146 pKa = 4.34NEE148 pKa = 4.55SDD150 pKa = 3.73EE151 pKa = 4.41TDD153 pKa = 3.51DD154 pKa = 6.43RR155 pKa = 11.84LDD157 pKa = 3.22TWVQAIQKK165 pKa = 9.76AA166 pKa = 3.58
MM1 pKa = 7.72KK2 pKa = 9.62NTGLYY7 pKa = 9.72FGSSTGHH14 pKa = 5.65VEE16 pKa = 4.83GIADD20 pKa = 4.58KK21 pKa = 10.81VAQLLGIDD29 pKa = 3.73AANVHH34 pKa = 5.87NVADD38 pKa = 4.16ATPEE42 pKa = 3.88SALAYY47 pKa = 10.07DD48 pKa = 4.23LLIFGSSTWGYY59 pKa = 11.66GDD61 pKa = 5.61LQDD64 pKa = 4.25DD65 pKa = 4.12WEE67 pKa = 4.79GFLPKK72 pKa = 10.14LVKK75 pKa = 9.92QDD77 pKa = 4.06LSGKK81 pKa = 8.56QFAVFGCGDD90 pKa = 3.45SSSYY94 pKa = 11.05PDD96 pKa = 3.71TFCDD100 pKa = 3.4AMAEE104 pKa = 4.0IAEE107 pKa = 4.33QVTAAGATLVGQIPAEE123 pKa = 4.61GYY125 pKa = 9.35SYY127 pKa = 10.86EE128 pKa = 4.05ATRR131 pKa = 11.84CEE133 pKa = 4.32VDD135 pKa = 3.35GQLIGCCLDD144 pKa = 3.73EE145 pKa = 6.29DD146 pKa = 4.34NEE148 pKa = 4.55SDD150 pKa = 3.73EE151 pKa = 4.41TDD153 pKa = 3.51DD154 pKa = 6.43RR155 pKa = 11.84LDD157 pKa = 3.22TWVQAIQKK165 pKa = 9.76AA166 pKa = 3.58
Molecular weight: 17.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4KMV5|F4KMV5_PORAD Uncharacterized protein OS=Porphyromonas asaccharolytica (strain ATCC 25260 / DSM 20707 / BCRC 10618 / JCM 6326 / LMG 13178 / VPI 4198) OX=879243 GN=Poras_1399 PE=4 SV=1
MM1 pKa = 6.94GQKK4 pKa = 9.78INPIANRR11 pKa = 11.84LGYY14 pKa = 9.7IRR16 pKa = 11.84GWDD19 pKa = 3.72SNWYY23 pKa = 9.83GGRR26 pKa = 11.84NYY28 pKa = 11.08GDD30 pKa = 3.52TLVEE34 pKa = 4.17DD35 pKa = 3.87DD36 pKa = 5.19RR37 pKa = 11.84IRR39 pKa = 11.84KK40 pKa = 8.65YY41 pKa = 11.04LHH43 pKa = 6.04TRR45 pKa = 11.84LSGALVSRR53 pKa = 11.84IIIEE57 pKa = 4.06RR58 pKa = 11.84ALKK61 pKa = 10.65VITITICTARR71 pKa = 11.84PGMIVGKK78 pKa = 10.44GGQNVEE84 pKa = 3.99NLKK87 pKa = 11.07AEE89 pKa = 4.3LKK91 pKa = 10.68KK92 pKa = 9.74VTKK95 pKa = 10.43ADD97 pKa = 3.85NIQINIYY104 pKa = 7.16EE105 pKa = 4.08VRR107 pKa = 11.84KK108 pKa = 9.77PEE110 pKa = 3.61VDD112 pKa = 3.31AVIVANDD119 pKa = 3.45IARR122 pKa = 11.84KK123 pKa = 9.91LEE125 pKa = 3.41GRR127 pKa = 11.84MSYY130 pKa = 10.47RR131 pKa = 11.84RR132 pKa = 11.84TVKK135 pKa = 9.89MAIANAMRR143 pKa = 11.84AGAEE147 pKa = 4.36GIKK150 pKa = 9.94IQLSGRR156 pKa = 11.84LNNAEE161 pKa = 4.13MARR164 pKa = 11.84SEE166 pKa = 4.25LYY168 pKa = 10.6KK169 pKa = 10.53EE170 pKa = 4.32GRR172 pKa = 11.84TPLHH176 pKa = 5.9TLRR179 pKa = 11.84ADD181 pKa = 2.9IDD183 pKa = 4.02YY184 pKa = 10.29AQTGALTKK192 pKa = 10.73VGMIGVKK199 pKa = 8.65VWIMNGEE206 pKa = 4.09VYY208 pKa = 9.88EE209 pKa = 4.43RR210 pKa = 11.84RR211 pKa = 11.84DD212 pKa = 3.53LAPDD216 pKa = 3.84LSQPRR221 pKa = 11.84RR222 pKa = 11.84QGGRR226 pKa = 11.84RR227 pKa = 11.84GGDD230 pKa = 2.96SRR232 pKa = 11.84GEE234 pKa = 3.56RR235 pKa = 11.84RR236 pKa = 11.84GFRR239 pKa = 11.84RR240 pKa = 11.84GGRR243 pKa = 11.84NRR245 pKa = 11.84SNRR248 pKa = 3.39
MM1 pKa = 6.94GQKK4 pKa = 9.78INPIANRR11 pKa = 11.84LGYY14 pKa = 9.7IRR16 pKa = 11.84GWDD19 pKa = 3.72SNWYY23 pKa = 9.83GGRR26 pKa = 11.84NYY28 pKa = 11.08GDD30 pKa = 3.52TLVEE34 pKa = 4.17DD35 pKa = 3.87DD36 pKa = 5.19RR37 pKa = 11.84IRR39 pKa = 11.84KK40 pKa = 8.65YY41 pKa = 11.04LHH43 pKa = 6.04TRR45 pKa = 11.84LSGALVSRR53 pKa = 11.84IIIEE57 pKa = 4.06RR58 pKa = 11.84ALKK61 pKa = 10.65VITITICTARR71 pKa = 11.84PGMIVGKK78 pKa = 10.44GGQNVEE84 pKa = 3.99NLKK87 pKa = 11.07AEE89 pKa = 4.3LKK91 pKa = 10.68KK92 pKa = 9.74VTKK95 pKa = 10.43ADD97 pKa = 3.85NIQINIYY104 pKa = 7.16EE105 pKa = 4.08VRR107 pKa = 11.84KK108 pKa = 9.77PEE110 pKa = 3.61VDD112 pKa = 3.31AVIVANDD119 pKa = 3.45IARR122 pKa = 11.84KK123 pKa = 9.91LEE125 pKa = 3.41GRR127 pKa = 11.84MSYY130 pKa = 10.47RR131 pKa = 11.84RR132 pKa = 11.84TVKK135 pKa = 9.89MAIANAMRR143 pKa = 11.84AGAEE147 pKa = 4.36GIKK150 pKa = 9.94IQLSGRR156 pKa = 11.84LNNAEE161 pKa = 4.13MARR164 pKa = 11.84SEE166 pKa = 4.25LYY168 pKa = 10.6KK169 pKa = 10.53EE170 pKa = 4.32GRR172 pKa = 11.84TPLHH176 pKa = 5.9TLRR179 pKa = 11.84ADD181 pKa = 2.9IDD183 pKa = 4.02YY184 pKa = 10.29AQTGALTKK192 pKa = 10.73VGMIGVKK199 pKa = 8.65VWIMNGEE206 pKa = 4.09VYY208 pKa = 9.88EE209 pKa = 4.43RR210 pKa = 11.84RR211 pKa = 11.84DD212 pKa = 3.53LAPDD216 pKa = 3.84LSQPRR221 pKa = 11.84RR222 pKa = 11.84QGGRR226 pKa = 11.84RR227 pKa = 11.84GGDD230 pKa = 2.96SRR232 pKa = 11.84GEE234 pKa = 3.56RR235 pKa = 11.84RR236 pKa = 11.84GFRR239 pKa = 11.84RR240 pKa = 11.84GGRR243 pKa = 11.84NRR245 pKa = 11.84SNRR248 pKa = 3.39
Molecular weight: 27.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
625607 |
32 |
2517 |
374.2 |
41.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.093 ± 0.052 | 1.131 ± 0.025 |
5.439 ± 0.031 | 6.523 ± 0.055 |
3.429 ± 0.033 | 6.805 ± 0.048 |
2.213 ± 0.028 | 6.274 ± 0.043 |
4.698 ± 0.047 | 10.618 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.451 ± 0.027 | 3.523 ± 0.046 |
4.436 ± 0.033 | 4.38 ± 0.037 |
5.515 ± 0.056 | 6.421 ± 0.054 |
6.274 ± 0.056 | 6.498 ± 0.043 |
1.09 ± 0.02 | 4.187 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
