
Pseudallescheria apiosperma (Scedosporium apiospermum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomyc
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
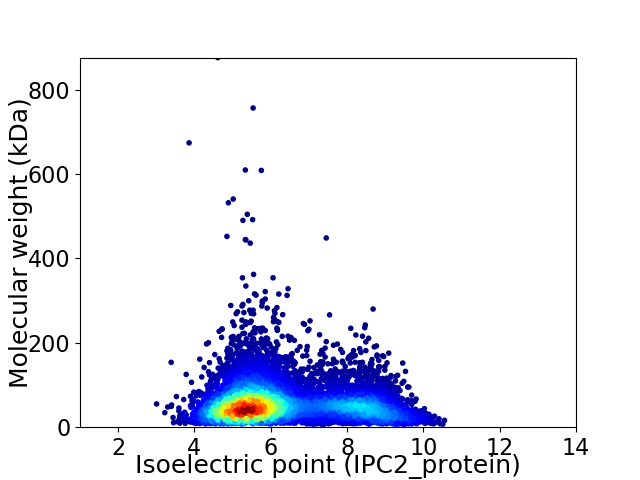
Virtual 2D-PAGE plot for 8375 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084G3H2|A0A084G3H2_PSEDA Fumarylacetoacetase OS=Pseudallescheria apiosperma OX=563466 GN=SAPIO_CDS6573 PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 10.48SQFLSLYY9 pKa = 9.83GLAALASLGVGAAARR24 pKa = 11.84TAEE27 pKa = 4.13PTVVLSARR35 pKa = 11.84SFKK38 pKa = 11.03HH39 pKa = 4.38EE40 pKa = 4.2ACWTDD45 pKa = 3.24EE46 pKa = 4.41DD47 pKa = 4.63ALSGPGFSSPSMTVRR62 pKa = 11.84WCARR66 pKa = 11.84FCRR69 pKa = 11.84GYY71 pKa = 11.02EE72 pKa = 3.92YY73 pKa = 10.77FGLANGDD80 pKa = 3.41GRR82 pKa = 11.84GSTRR86 pKa = 11.84LSLYY90 pKa = 9.47KK91 pKa = 10.45AKK93 pKa = 10.72EE94 pKa = 3.92GGTCHH99 pKa = 5.95VRR101 pKa = 11.84PSSTRR106 pKa = 11.84AHH108 pKa = 6.15SSEE111 pKa = 4.29SEE113 pKa = 4.06IMTEE117 pKa = 4.07PATSTSASEE126 pKa = 4.1LTSATIEE133 pKa = 4.25SAQTTEE139 pKa = 4.77SITEE143 pKa = 4.09SASATEE149 pKa = 4.53SEE151 pKa = 4.94SPTSTEE157 pKa = 3.86EE158 pKa = 3.97ASEE161 pKa = 4.33STSEE165 pKa = 4.26SAIEE169 pKa = 4.2SDD171 pKa = 3.48TAPVTEE177 pKa = 5.07SEE179 pKa = 4.47TSSATLTQSQPEE191 pKa = 4.3TTFDD195 pKa = 3.5VTTSTSEE202 pKa = 4.12SATEE206 pKa = 3.57TDD208 pKa = 3.17AASTSEE214 pKa = 4.15STVEE218 pKa = 4.1SSSEE222 pKa = 4.06SATEE226 pKa = 4.04LSTEE230 pKa = 4.28SEE232 pKa = 4.4PEE234 pKa = 3.82PTTTAEE240 pKa = 4.33LEE242 pKa = 4.37STTTEE247 pKa = 3.98DD248 pKa = 3.35AAPATTSDD256 pKa = 4.15SEE258 pKa = 4.5SSTTEE263 pKa = 3.79EE264 pKa = 4.54PEE266 pKa = 4.17TTVTSDD272 pKa = 4.45AEE274 pKa = 4.35TTTTKK279 pKa = 10.56EE280 pKa = 4.15PEE282 pKa = 4.44TTTVTPDD289 pKa = 3.02VEE291 pKa = 4.54TTTAEE296 pKa = 4.14EE297 pKa = 4.22PVATTTEE304 pKa = 4.2EE305 pKa = 4.52PEE307 pKa = 4.16TTSAAEE313 pKa = 4.0TTTTEE318 pKa = 3.97EE319 pKa = 4.45SEE321 pKa = 4.36TTSASDD327 pKa = 3.06VDD329 pKa = 3.95TTTTEE334 pKa = 4.1EE335 pKa = 4.18ASTTTTEE342 pKa = 4.15DD343 pKa = 3.9PEE345 pKa = 4.61TKK347 pKa = 9.98TIEE350 pKa = 4.25DD351 pKa = 3.91TSTSTTEE358 pKa = 4.35TPTSTPTPCIDD369 pKa = 4.05CQNLPTGNWLLASTSTGSGILEE391 pKa = 4.15LRR393 pKa = 11.84YY394 pKa = 10.47DD395 pKa = 3.72SAGGYY400 pKa = 8.67YY401 pKa = 9.7YY402 pKa = 10.99VYY404 pKa = 10.37NEE406 pKa = 4.84CPASHH411 pKa = 6.78GGYY414 pKa = 7.54CTVYY418 pKa = 8.03IAQVISVEE426 pKa = 4.07AGITYY431 pKa = 10.38DD432 pKa = 4.42FAIQYY437 pKa = 10.42LMSNVRR443 pKa = 11.84NQANFLQISISTLPSRR459 pKa = 11.84TRR461 pKa = 11.84IFTEE465 pKa = 4.26YY466 pKa = 10.28FSSGSTNGWTDD477 pKa = 3.62FRR479 pKa = 11.84TSPFTPTVSGDD490 pKa = 3.41VLITVNWLNDD500 pKa = 3.72PNNALVLIRR509 pKa = 11.84NIVMQPTEE517 pKa = 4.07CRR519 pKa = 11.84NPLSEE524 pKa = 4.4LTCNTDD530 pKa = 2.96VEE532 pKa = 4.72VQVVV536 pKa = 3.14
MM1 pKa = 7.71KK2 pKa = 10.48SQFLSLYY9 pKa = 9.83GLAALASLGVGAAARR24 pKa = 11.84TAEE27 pKa = 4.13PTVVLSARR35 pKa = 11.84SFKK38 pKa = 11.03HH39 pKa = 4.38EE40 pKa = 4.2ACWTDD45 pKa = 3.24EE46 pKa = 4.41DD47 pKa = 4.63ALSGPGFSSPSMTVRR62 pKa = 11.84WCARR66 pKa = 11.84FCRR69 pKa = 11.84GYY71 pKa = 11.02EE72 pKa = 3.92YY73 pKa = 10.77FGLANGDD80 pKa = 3.41GRR82 pKa = 11.84GSTRR86 pKa = 11.84LSLYY90 pKa = 9.47KK91 pKa = 10.45AKK93 pKa = 10.72EE94 pKa = 3.92GGTCHH99 pKa = 5.95VRR101 pKa = 11.84PSSTRR106 pKa = 11.84AHH108 pKa = 6.15SSEE111 pKa = 4.29SEE113 pKa = 4.06IMTEE117 pKa = 4.07PATSTSASEE126 pKa = 4.1LTSATIEE133 pKa = 4.25SAQTTEE139 pKa = 4.77SITEE143 pKa = 4.09SASATEE149 pKa = 4.53SEE151 pKa = 4.94SPTSTEE157 pKa = 3.86EE158 pKa = 3.97ASEE161 pKa = 4.33STSEE165 pKa = 4.26SAIEE169 pKa = 4.2SDD171 pKa = 3.48TAPVTEE177 pKa = 5.07SEE179 pKa = 4.47TSSATLTQSQPEE191 pKa = 4.3TTFDD195 pKa = 3.5VTTSTSEE202 pKa = 4.12SATEE206 pKa = 3.57TDD208 pKa = 3.17AASTSEE214 pKa = 4.15STVEE218 pKa = 4.1SSSEE222 pKa = 4.06SATEE226 pKa = 4.04LSTEE230 pKa = 4.28SEE232 pKa = 4.4PEE234 pKa = 3.82PTTTAEE240 pKa = 4.33LEE242 pKa = 4.37STTTEE247 pKa = 3.98DD248 pKa = 3.35AAPATTSDD256 pKa = 4.15SEE258 pKa = 4.5SSTTEE263 pKa = 3.79EE264 pKa = 4.54PEE266 pKa = 4.17TTVTSDD272 pKa = 4.45AEE274 pKa = 4.35TTTTKK279 pKa = 10.56EE280 pKa = 4.15PEE282 pKa = 4.44TTTVTPDD289 pKa = 3.02VEE291 pKa = 4.54TTTAEE296 pKa = 4.14EE297 pKa = 4.22PVATTTEE304 pKa = 4.2EE305 pKa = 4.52PEE307 pKa = 4.16TTSAAEE313 pKa = 4.0TTTTEE318 pKa = 3.97EE319 pKa = 4.45SEE321 pKa = 4.36TTSASDD327 pKa = 3.06VDD329 pKa = 3.95TTTTEE334 pKa = 4.1EE335 pKa = 4.18ASTTTTEE342 pKa = 4.15DD343 pKa = 3.9PEE345 pKa = 4.61TKK347 pKa = 9.98TIEE350 pKa = 4.25DD351 pKa = 3.91TSTSTTEE358 pKa = 4.35TPTSTPTPCIDD369 pKa = 4.05CQNLPTGNWLLASTSTGSGILEE391 pKa = 4.15LRR393 pKa = 11.84YY394 pKa = 10.47DD395 pKa = 3.72SAGGYY400 pKa = 8.67YY401 pKa = 9.7YY402 pKa = 10.99VYY404 pKa = 10.37NEE406 pKa = 4.84CPASHH411 pKa = 6.78GGYY414 pKa = 7.54CTVYY418 pKa = 8.03IAQVISVEE426 pKa = 4.07AGITYY431 pKa = 10.38DD432 pKa = 4.42FAIQYY437 pKa = 10.42LMSNVRR443 pKa = 11.84NQANFLQISISTLPSRR459 pKa = 11.84TRR461 pKa = 11.84IFTEE465 pKa = 4.26YY466 pKa = 10.28FSSGSTNGWTDD477 pKa = 3.62FRR479 pKa = 11.84TSPFTPTVSGDD490 pKa = 3.41VLITVNWLNDD500 pKa = 3.72PNNALVLIRR509 pKa = 11.84NIVMQPTEE517 pKa = 4.07CRR519 pKa = 11.84NPLSEE524 pKa = 4.4LTCNTDD530 pKa = 2.96VEE532 pKa = 4.72VQVVV536 pKa = 3.14
Molecular weight: 56.85 kDa
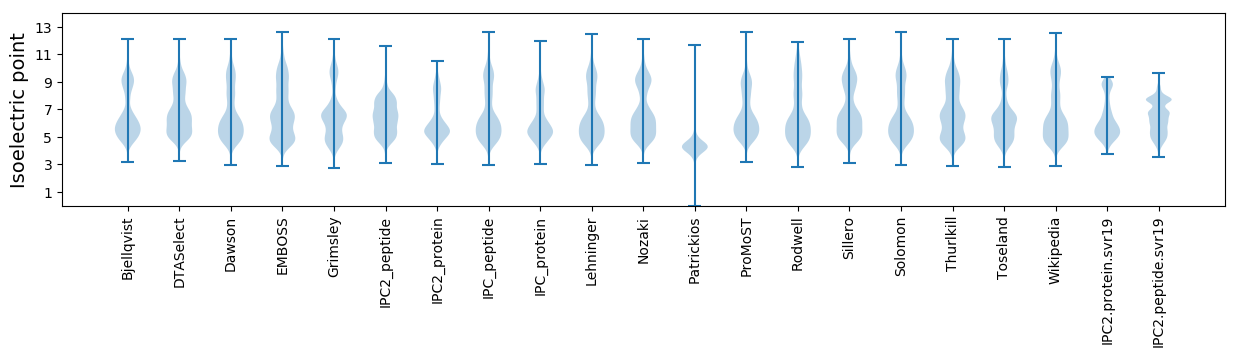
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A084GDE5|A0A084GDE5_PSEDA Ribosome assembly protein 1 OS=Pseudallescheria apiosperma OX=563466 GN=SAPIO_CDS2175 PE=4 SV=1
MM1 pKa = 7.36SALATSQDD9 pKa = 3.58TRR11 pKa = 11.84CTPNISNDD19 pKa = 3.56AFCMTTAKK27 pKa = 10.06RR28 pKa = 11.84HH29 pKa = 5.62RR30 pKa = 11.84PSNTIRR36 pKa = 11.84PRR38 pKa = 11.84IAWADD43 pKa = 3.49TTMTTNPLMVIHH55 pKa = 6.81PPPYY59 pKa = 10.0LNNSAEE65 pKa = 4.15ICRR68 pKa = 11.84SSRR71 pKa = 11.84NDD73 pKa = 2.92IAGIRR78 pKa = 11.84PPRR81 pKa = 11.84RR82 pKa = 11.84GSMVMALVFTPRR94 pKa = 11.84DD95 pKa = 3.29KK96 pKa = 10.96EE97 pKa = 4.07RR98 pKa = 11.84RR99 pKa = 11.84DD100 pKa = 3.43EE101 pKa = 4.12KK102 pKa = 11.41
MM1 pKa = 7.36SALATSQDD9 pKa = 3.58TRR11 pKa = 11.84CTPNISNDD19 pKa = 3.56AFCMTTAKK27 pKa = 10.06RR28 pKa = 11.84HH29 pKa = 5.62RR30 pKa = 11.84PSNTIRR36 pKa = 11.84PRR38 pKa = 11.84IAWADD43 pKa = 3.49TTMTTNPLMVIHH55 pKa = 6.81PPPYY59 pKa = 10.0LNNSAEE65 pKa = 4.15ICRR68 pKa = 11.84SSRR71 pKa = 11.84NDD73 pKa = 2.92IAGIRR78 pKa = 11.84PPRR81 pKa = 11.84RR82 pKa = 11.84GSMVMALVFTPRR94 pKa = 11.84DD95 pKa = 3.29KK96 pKa = 10.96EE97 pKa = 4.07RR98 pKa = 11.84RR99 pKa = 11.84DD100 pKa = 3.43EE101 pKa = 4.12KK102 pKa = 11.41
Molecular weight: 11.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4573252 |
66 |
8042 |
546.1 |
60.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.814 ± 0.027 | 1.211 ± 0.009 |
5.854 ± 0.02 | 6.395 ± 0.034 |
3.739 ± 0.018 | 7.095 ± 0.022 |
2.276 ± 0.011 | 4.951 ± 0.016 |
4.78 ± 0.022 | 8.964 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.082 ± 0.009 | 3.533 ± 0.015 |
6.19 ± 0.033 | 3.763 ± 0.02 |
6.166 ± 0.02 | 7.866 ± 0.027 |
5.842 ± 0.016 | 6.292 ± 0.019 |
1.47 ± 0.01 | 2.718 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
