
Helicobacter phage UKEN32U
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Schmidvirus; unclassified Schmidvirus
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
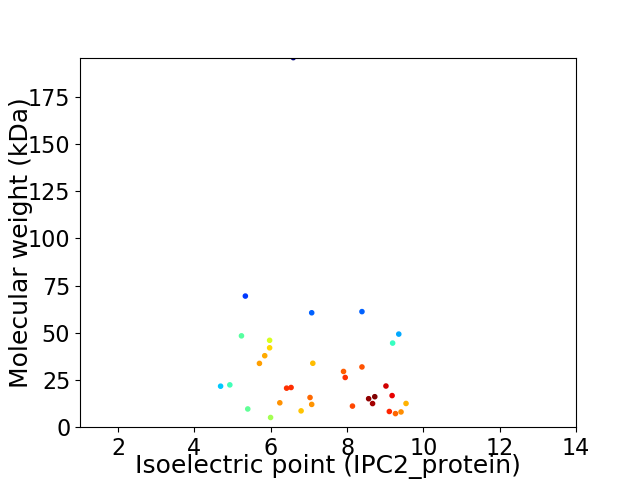
Virtual 2D-PAGE plot for 35 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S5RH61|A0A1S5RH61_9CAUD Mitogen-activated protein kinase 1 OS=Helicobacter phage UKEN32U OX=1852688 PE=4 SV=1
MM1 pKa = 7.66GIKK4 pKa = 9.82EE5 pKa = 4.23KK6 pKa = 10.55EE7 pKa = 3.99IEE9 pKa = 4.18LEE11 pKa = 3.99TLKK14 pKa = 11.01RR15 pKa = 11.84EE16 pKa = 3.99IAQAEE21 pKa = 4.06ASLEE25 pKa = 3.89QDD27 pKa = 4.59FIKK30 pKa = 10.93HH31 pKa = 5.17MVDD34 pKa = 2.85KK35 pKa = 10.51TSEE38 pKa = 4.11KK39 pKa = 10.83VEE41 pKa = 4.46DD42 pKa = 3.98LFFSNKK48 pKa = 8.85PEE50 pKa = 4.29FYY52 pKa = 10.01RR53 pKa = 11.84FVFNEE58 pKa = 3.46QNKK61 pKa = 8.73YY62 pKa = 10.52LRR64 pKa = 11.84EE65 pKa = 3.98KK66 pKa = 10.29LTDD69 pKa = 3.41KK70 pKa = 10.83VGRR73 pKa = 11.84AMDD76 pKa = 4.55LSDD79 pKa = 5.66EE80 pKa = 4.18IQNDD84 pKa = 2.98KK85 pKa = 10.97DD86 pKa = 3.56AEE88 pKa = 4.21EE89 pKa = 4.15IEE91 pKa = 4.13KK92 pKa = 11.03DD93 pKa = 3.3KK94 pKa = 11.44EE95 pKa = 4.14AFLKK99 pKa = 10.14KK100 pKa = 10.06HH101 pKa = 6.08PEE103 pKa = 3.38IDD105 pKa = 3.74FNEE108 pKa = 3.95LLEE111 pKa = 4.62FYY113 pKa = 10.85NEE115 pKa = 4.02EE116 pKa = 3.77VPNRR120 pKa = 11.84IKK122 pKa = 10.86KK123 pKa = 10.42QIDD126 pKa = 3.39KK127 pKa = 11.12LEE129 pKa = 4.21GVAFFEE135 pKa = 5.24AVLDD139 pKa = 3.87YY140 pKa = 11.19FNALNAKK147 pKa = 9.33EE148 pKa = 4.37EE149 pKa = 4.21EE150 pKa = 4.46PKK152 pKa = 10.95NEE154 pKa = 4.05AKK156 pKa = 10.59EE157 pKa = 4.24EE158 pKa = 3.97EE159 pKa = 4.33SQLPKK164 pKa = 10.37EE165 pKa = 4.03ALGNGVSGVGNASNEE180 pKa = 4.31NIMTRR185 pKa = 11.84YY186 pKa = 9.53
MM1 pKa = 7.66GIKK4 pKa = 9.82EE5 pKa = 4.23KK6 pKa = 10.55EE7 pKa = 3.99IEE9 pKa = 4.18LEE11 pKa = 3.99TLKK14 pKa = 11.01RR15 pKa = 11.84EE16 pKa = 3.99IAQAEE21 pKa = 4.06ASLEE25 pKa = 3.89QDD27 pKa = 4.59FIKK30 pKa = 10.93HH31 pKa = 5.17MVDD34 pKa = 2.85KK35 pKa = 10.51TSEE38 pKa = 4.11KK39 pKa = 10.83VEE41 pKa = 4.46DD42 pKa = 3.98LFFSNKK48 pKa = 8.85PEE50 pKa = 4.29FYY52 pKa = 10.01RR53 pKa = 11.84FVFNEE58 pKa = 3.46QNKK61 pKa = 8.73YY62 pKa = 10.52LRR64 pKa = 11.84EE65 pKa = 3.98KK66 pKa = 10.29LTDD69 pKa = 3.41KK70 pKa = 10.83VGRR73 pKa = 11.84AMDD76 pKa = 4.55LSDD79 pKa = 5.66EE80 pKa = 4.18IQNDD84 pKa = 2.98KK85 pKa = 10.97DD86 pKa = 3.56AEE88 pKa = 4.21EE89 pKa = 4.15IEE91 pKa = 4.13KK92 pKa = 11.03DD93 pKa = 3.3KK94 pKa = 11.44EE95 pKa = 4.14AFLKK99 pKa = 10.14KK100 pKa = 10.06HH101 pKa = 6.08PEE103 pKa = 3.38IDD105 pKa = 3.74FNEE108 pKa = 3.95LLEE111 pKa = 4.62FYY113 pKa = 10.85NEE115 pKa = 4.02EE116 pKa = 3.77VPNRR120 pKa = 11.84IKK122 pKa = 10.86KK123 pKa = 10.42QIDD126 pKa = 3.39KK127 pKa = 11.12LEE129 pKa = 4.21GVAFFEE135 pKa = 5.24AVLDD139 pKa = 3.87YY140 pKa = 11.19FNALNAKK147 pKa = 9.33EE148 pKa = 4.37EE149 pKa = 4.21EE150 pKa = 4.46PKK152 pKa = 10.95NEE154 pKa = 4.05AKK156 pKa = 10.59EE157 pKa = 4.24EE158 pKa = 3.97EE159 pKa = 4.33SQLPKK164 pKa = 10.37EE165 pKa = 4.03ALGNGVSGVGNASNEE180 pKa = 4.31NIMTRR185 pKa = 11.84YY186 pKa = 9.53
Molecular weight: 21.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S5RH52|A0A1S5RH52_9CAUD Putative terminase OS=Helicobacter phage UKEN32U OX=1852688 PE=4 SV=1
MM1 pKa = 7.44LNAIKK6 pKa = 10.31FRR8 pKa = 11.84IYY10 pKa = 10.58PNAQQKK16 pKa = 9.41EE17 pKa = 4.62LISKK21 pKa = 10.01HH22 pKa = 5.57FGCSRR27 pKa = 11.84VVYY30 pKa = 10.47NYY32 pKa = 10.55FLDD35 pKa = 3.97YY36 pKa = 10.43RR37 pKa = 11.84QKK39 pKa = 10.83QYY41 pKa = 11.81AKK43 pKa = 10.14GIKK46 pKa = 8.01EE47 pKa = 4.53TYY49 pKa = 8.68FTMQKK54 pKa = 10.2VLTQIKK60 pKa = 9.44RR61 pKa = 11.84QEE63 pKa = 3.97KK64 pKa = 8.17YY65 pKa = 10.47HH66 pKa = 6.16YY67 pKa = 10.84LNEE70 pKa = 4.56CNSQSLQMALRR81 pKa = 11.84QLVSAYY87 pKa = 10.93DD88 pKa = 3.42NFFSKK93 pKa = 10.5RR94 pKa = 11.84ARR96 pKa = 11.84YY97 pKa = 8.55PKK99 pKa = 10.37FKK101 pKa = 10.41SKK103 pKa = 10.76KK104 pKa = 7.81NAKK107 pKa = 9.36QSFAIPQNIEE117 pKa = 3.39IKK119 pKa = 11.0AEE121 pKa = 4.12TKK123 pKa = 10.19TIALPKK129 pKa = 9.84FKK131 pKa = 10.72EE132 pKa = 4.62GIKK135 pKa = 10.62AKK137 pKa = 9.61LHH139 pKa = 6.41RR140 pKa = 11.84DD141 pKa = 3.43LPKK144 pKa = 11.01DD145 pKa = 3.71SVIKK149 pKa = 10.32QAFISCIADD158 pKa = 3.12QYY160 pKa = 10.88FCSLSYY166 pKa = 8.8EE167 pKa = 4.33TKK169 pKa = 10.58EE170 pKa = 4.6PIPKK174 pKa = 8.77PTIIKK179 pKa = 10.01KK180 pKa = 10.46AVGLDD185 pKa = 3.03MGLRR189 pKa = 11.84TLIVTSDD196 pKa = 3.47KK197 pKa = 10.91IEE199 pKa = 4.06YY200 pKa = 8.68PHH202 pKa = 7.5IRR204 pKa = 11.84FYY206 pKa = 11.37QKK208 pKa = 10.84LEE210 pKa = 4.0KK211 pKa = 10.51KK212 pKa = 8.72LTKK215 pKa = 9.84AQRR218 pKa = 11.84RR219 pKa = 11.84LSKK222 pKa = 10.28KK223 pKa = 10.36VKK225 pKa = 10.08GSNNRR230 pKa = 11.84KK231 pKa = 8.63KK232 pKa = 9.9QAKK235 pKa = 9.04KK236 pKa = 10.2VSRR239 pKa = 11.84LHH241 pKa = 6.25LACSNTRR248 pKa = 11.84DD249 pKa = 4.0DD250 pKa = 4.33YY251 pKa = 11.62LHH253 pKa = 6.99KK254 pKa = 10.44ISNEE258 pKa = 3.23ITNQYY263 pKa = 10.97DD264 pKa = 4.17LIGVEE269 pKa = 4.16TLNIKK274 pKa = 10.64ALMKK278 pKa = 9.63TYY280 pKa = 10.22HH281 pKa = 6.41SKK283 pKa = 10.96SLANASWGKK292 pKa = 9.22FLAMLEE298 pKa = 4.21YY299 pKa = 10.38KK300 pKa = 10.32AQRR303 pKa = 11.84KK304 pKa = 9.03GKK306 pKa = 7.34TLLRR310 pKa = 11.84IDD312 pKa = 3.95RR313 pKa = 11.84FFPSSQLCSYY323 pKa = 10.69CGVNTGKK330 pKa = 9.97KK331 pKa = 9.39HH332 pKa = 5.53EE333 pKa = 5.31SITKK337 pKa = 7.35FTCPHH342 pKa = 6.91CKK344 pKa = 7.53TTHH347 pKa = 5.66HH348 pKa = 7.02RR349 pKa = 11.84DD350 pKa = 3.21YY351 pKa = 10.78NASVNIRR358 pKa = 11.84NYY360 pKa = 10.82ALGMLDD366 pKa = 4.8DD367 pKa = 3.81RR368 pKa = 11.84HH369 pKa = 6.14KK370 pKa = 11.43VKK372 pKa = 10.0TDD374 pKa = 3.04KK375 pKa = 11.22ARR377 pKa = 11.84VGIIRR382 pKa = 11.84SDD384 pKa = 3.75YY385 pKa = 7.91THH387 pKa = 6.78HH388 pKa = 6.81TNEE391 pKa = 4.28CVKK394 pKa = 11.0ACGASSNGVSSKK406 pKa = 10.83YY407 pKa = 11.2GNILDD412 pKa = 4.17LASYY416 pKa = 10.91GAMKK420 pKa = 10.18QEE422 pKa = 4.59KK423 pKa = 9.54AQSLL427 pKa = 3.89
MM1 pKa = 7.44LNAIKK6 pKa = 10.31FRR8 pKa = 11.84IYY10 pKa = 10.58PNAQQKK16 pKa = 9.41EE17 pKa = 4.62LISKK21 pKa = 10.01HH22 pKa = 5.57FGCSRR27 pKa = 11.84VVYY30 pKa = 10.47NYY32 pKa = 10.55FLDD35 pKa = 3.97YY36 pKa = 10.43RR37 pKa = 11.84QKK39 pKa = 10.83QYY41 pKa = 11.81AKK43 pKa = 10.14GIKK46 pKa = 8.01EE47 pKa = 4.53TYY49 pKa = 8.68FTMQKK54 pKa = 10.2VLTQIKK60 pKa = 9.44RR61 pKa = 11.84QEE63 pKa = 3.97KK64 pKa = 8.17YY65 pKa = 10.47HH66 pKa = 6.16YY67 pKa = 10.84LNEE70 pKa = 4.56CNSQSLQMALRR81 pKa = 11.84QLVSAYY87 pKa = 10.93DD88 pKa = 3.42NFFSKK93 pKa = 10.5RR94 pKa = 11.84ARR96 pKa = 11.84YY97 pKa = 8.55PKK99 pKa = 10.37FKK101 pKa = 10.41SKK103 pKa = 10.76KK104 pKa = 7.81NAKK107 pKa = 9.36QSFAIPQNIEE117 pKa = 3.39IKK119 pKa = 11.0AEE121 pKa = 4.12TKK123 pKa = 10.19TIALPKK129 pKa = 9.84FKK131 pKa = 10.72EE132 pKa = 4.62GIKK135 pKa = 10.62AKK137 pKa = 9.61LHH139 pKa = 6.41RR140 pKa = 11.84DD141 pKa = 3.43LPKK144 pKa = 11.01DD145 pKa = 3.71SVIKK149 pKa = 10.32QAFISCIADD158 pKa = 3.12QYY160 pKa = 10.88FCSLSYY166 pKa = 8.8EE167 pKa = 4.33TKK169 pKa = 10.58EE170 pKa = 4.6PIPKK174 pKa = 8.77PTIIKK179 pKa = 10.01KK180 pKa = 10.46AVGLDD185 pKa = 3.03MGLRR189 pKa = 11.84TLIVTSDD196 pKa = 3.47KK197 pKa = 10.91IEE199 pKa = 4.06YY200 pKa = 8.68PHH202 pKa = 7.5IRR204 pKa = 11.84FYY206 pKa = 11.37QKK208 pKa = 10.84LEE210 pKa = 4.0KK211 pKa = 10.51KK212 pKa = 8.72LTKK215 pKa = 9.84AQRR218 pKa = 11.84RR219 pKa = 11.84LSKK222 pKa = 10.28KK223 pKa = 10.36VKK225 pKa = 10.08GSNNRR230 pKa = 11.84KK231 pKa = 8.63KK232 pKa = 9.9QAKK235 pKa = 9.04KK236 pKa = 10.2VSRR239 pKa = 11.84LHH241 pKa = 6.25LACSNTRR248 pKa = 11.84DD249 pKa = 4.0DD250 pKa = 4.33YY251 pKa = 11.62LHH253 pKa = 6.99KK254 pKa = 10.44ISNEE258 pKa = 3.23ITNQYY263 pKa = 10.97DD264 pKa = 4.17LIGVEE269 pKa = 4.16TLNIKK274 pKa = 10.64ALMKK278 pKa = 9.63TYY280 pKa = 10.22HH281 pKa = 6.41SKK283 pKa = 10.96SLANASWGKK292 pKa = 9.22FLAMLEE298 pKa = 4.21YY299 pKa = 10.38KK300 pKa = 10.32AQRR303 pKa = 11.84KK304 pKa = 9.03GKK306 pKa = 7.34TLLRR310 pKa = 11.84IDD312 pKa = 3.95RR313 pKa = 11.84FFPSSQLCSYY323 pKa = 10.69CGVNTGKK330 pKa = 9.97KK331 pKa = 9.39HH332 pKa = 5.53EE333 pKa = 5.31SITKK337 pKa = 7.35FTCPHH342 pKa = 6.91CKK344 pKa = 7.53TTHH347 pKa = 5.66HH348 pKa = 7.02RR349 pKa = 11.84DD350 pKa = 3.21YY351 pKa = 10.78NASVNIRR358 pKa = 11.84NYY360 pKa = 10.82ALGMLDD366 pKa = 4.8DD367 pKa = 3.81RR368 pKa = 11.84HH369 pKa = 6.14KK370 pKa = 11.43VKK372 pKa = 10.0TDD374 pKa = 3.04KK375 pKa = 11.22ARR377 pKa = 11.84VGIIRR382 pKa = 11.84SDD384 pKa = 3.75YY385 pKa = 7.91THH387 pKa = 6.78HH388 pKa = 6.81TNEE391 pKa = 4.28CVKK394 pKa = 11.0ACGASSNGVSSKK406 pKa = 10.83YY407 pKa = 11.2GNILDD412 pKa = 4.17LASYY416 pKa = 10.91GAMKK420 pKa = 10.18QEE422 pKa = 4.59KK423 pKa = 9.54AQSLL427 pKa = 3.89
Molecular weight: 49.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9506 |
48 |
1724 |
271.6 |
31.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.627 ± 0.347 | 0.799 ± 0.213 |
4.881 ± 0.49 | 8.405 ± 0.405 |
4.892 ± 0.295 | 4.176 ± 0.254 |
1.42 ± 0.186 | 7.122 ± 0.273 |
10.814 ± 0.484 | 10.793 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.704 ± 0.191 | 8.121 ± 0.479 |
2.356 ± 0.188 | 4.66 ± 0.392 |
3.766 ± 0.251 | 6.47 ± 0.271 |
4.86 ± 0.397 | 4.166 ± 0.36 |
0.442 ± 0.077 | 3.524 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
