
Pontibacter fetidus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Pontibacter
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
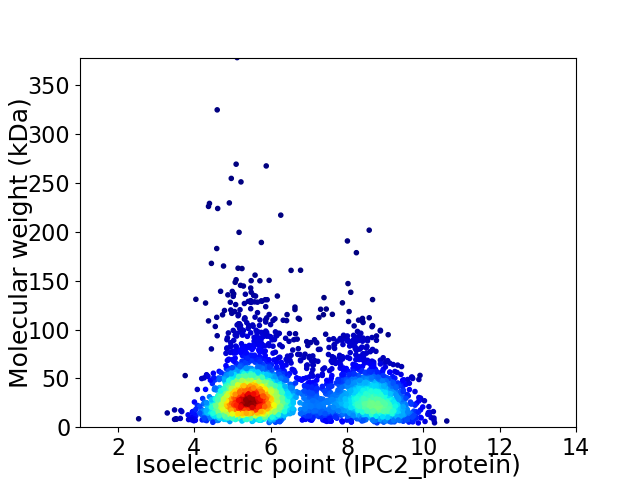
Virtual 2D-PAGE plot for 3463 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
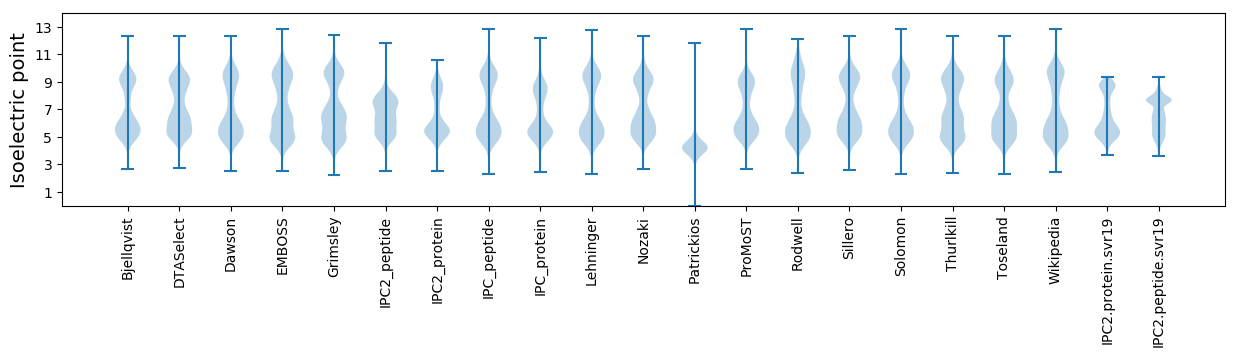
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B2H9T1|A0A6B2H9T1_9BACT Sigma-54-dependent Fis family transcriptional regulator OS=Pontibacter fetidus OX=2700082 GN=GWO68_13720 PE=4 SV=1
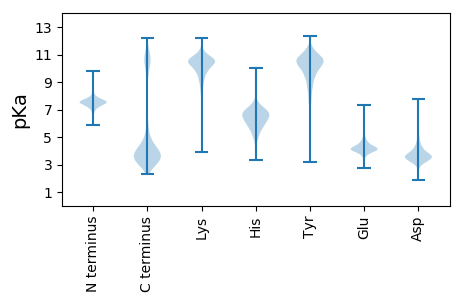
MM1 pKa = 7.61KK2 pKa = 10.46NFNNYY7 pKa = 6.9TSRR10 pKa = 11.84NSSILSKK17 pKa = 10.85QLLCSLLFGLLVFMPAVLHH36 pKa = 6.31ANYY39 pKa = 10.26VSLNKK44 pKa = 8.32TTEE47 pKa = 3.48IRR49 pKa = 11.84NTLLEE54 pKa = 4.0FDD56 pKa = 3.95NNKK59 pKa = 9.78VKK61 pKa = 10.79DD62 pKa = 4.07ILWSAIGITNDD73 pKa = 2.71VDD75 pKa = 3.64YY76 pKa = 11.47HH77 pKa = 6.55KK78 pKa = 10.65VTEE81 pKa = 4.14AKK83 pKa = 10.28FDD85 pKa = 3.83GGDD88 pKa = 3.38FSLDD92 pKa = 3.5FVASSPNTYY101 pKa = 10.57DD102 pKa = 3.02HH103 pKa = 6.36STGGGAFDD111 pKa = 4.11TRR113 pKa = 11.84DD114 pKa = 3.22VGQDD118 pKa = 2.91TGDD121 pKa = 4.76DD122 pKa = 3.25IVEE125 pKa = 4.25SLEE128 pKa = 4.89GGDD131 pKa = 4.03FTCGDD136 pKa = 3.18IVTFLTQIVVDD147 pKa = 4.14KK148 pKa = 10.86DD149 pKa = 3.57AVGSQTIDD157 pKa = 3.13LDD159 pKa = 4.16YY160 pKa = 11.49EE161 pKa = 4.22FLADD165 pKa = 3.49ATGQSGVALADD176 pKa = 3.29IVNVQINYY184 pKa = 9.49GAVSGGDD191 pKa = 3.66GVGSTDD197 pKa = 5.02SGIKK201 pKa = 10.16DD202 pKa = 3.68DD203 pKa = 4.62GGSVATLSNEE213 pKa = 4.2HH214 pKa = 6.02ITGDD218 pKa = 3.84GEE220 pKa = 4.64LFNDD224 pKa = 3.58NKK226 pKa = 10.95LVGTVTITDD235 pKa = 4.26LEE237 pKa = 4.21ASEE240 pKa = 4.18EE241 pKa = 4.41VIVRR245 pKa = 11.84VDD247 pKa = 3.07VRR249 pKa = 11.84IACDD253 pKa = 3.59PTSSPTGNLQAAITAALVIDD273 pKa = 4.84PEE275 pKa = 5.39DD276 pKa = 3.83DD277 pKa = 3.96VISVGNQTVPFKK289 pKa = 10.91NVNQIIFPTCAITPATAVCEE309 pKa = 4.07LATTSYY315 pKa = 9.61TASSDD320 pKa = 3.57VADD323 pKa = 3.92ATFVWSITGDD333 pKa = 3.58GTIVDD338 pKa = 4.11AASDD342 pKa = 3.77GTKK345 pKa = 10.14TIVASGGATSTSVSVLAGSAGSYY368 pKa = 8.33TLSVSISKK376 pKa = 8.97TGFGTQTCSQEE387 pKa = 3.56ITVNDD392 pKa = 4.01VPTVTAGADD401 pKa = 3.4QTVCAASPIQTLTATATVSTGATLVWYY428 pKa = 9.33DD429 pKa = 3.61AASGGNTVANPTLSSIGTATYY450 pKa = 9.77YY451 pKa = 10.99AQANLGDD458 pKa = 4.28CSSGRR463 pKa = 11.84VPVVLTINEE472 pKa = 4.54TPTVTAGADD481 pKa = 3.4QTVCAASPIQTLTATATVSTGATLVWYY508 pKa = 9.33DD509 pKa = 3.65AASGGNN515 pKa = 3.39
MM1 pKa = 7.61KK2 pKa = 10.46NFNNYY7 pKa = 6.9TSRR10 pKa = 11.84NSSILSKK17 pKa = 10.85QLLCSLLFGLLVFMPAVLHH36 pKa = 6.31ANYY39 pKa = 10.26VSLNKK44 pKa = 8.32TTEE47 pKa = 3.48IRR49 pKa = 11.84NTLLEE54 pKa = 4.0FDD56 pKa = 3.95NNKK59 pKa = 9.78VKK61 pKa = 10.79DD62 pKa = 4.07ILWSAIGITNDD73 pKa = 2.71VDD75 pKa = 3.64YY76 pKa = 11.47HH77 pKa = 6.55KK78 pKa = 10.65VTEE81 pKa = 4.14AKK83 pKa = 10.28FDD85 pKa = 3.83GGDD88 pKa = 3.38FSLDD92 pKa = 3.5FVASSPNTYY101 pKa = 10.57DD102 pKa = 3.02HH103 pKa = 6.36STGGGAFDD111 pKa = 4.11TRR113 pKa = 11.84DD114 pKa = 3.22VGQDD118 pKa = 2.91TGDD121 pKa = 4.76DD122 pKa = 3.25IVEE125 pKa = 4.25SLEE128 pKa = 4.89GGDD131 pKa = 4.03FTCGDD136 pKa = 3.18IVTFLTQIVVDD147 pKa = 4.14KK148 pKa = 10.86DD149 pKa = 3.57AVGSQTIDD157 pKa = 3.13LDD159 pKa = 4.16YY160 pKa = 11.49EE161 pKa = 4.22FLADD165 pKa = 3.49ATGQSGVALADD176 pKa = 3.29IVNVQINYY184 pKa = 9.49GAVSGGDD191 pKa = 3.66GVGSTDD197 pKa = 5.02SGIKK201 pKa = 10.16DD202 pKa = 3.68DD203 pKa = 4.62GGSVATLSNEE213 pKa = 4.2HH214 pKa = 6.02ITGDD218 pKa = 3.84GEE220 pKa = 4.64LFNDD224 pKa = 3.58NKK226 pKa = 10.95LVGTVTITDD235 pKa = 4.26LEE237 pKa = 4.21ASEE240 pKa = 4.18EE241 pKa = 4.41VIVRR245 pKa = 11.84VDD247 pKa = 3.07VRR249 pKa = 11.84IACDD253 pKa = 3.59PTSSPTGNLQAAITAALVIDD273 pKa = 4.84PEE275 pKa = 5.39DD276 pKa = 3.83DD277 pKa = 3.96VISVGNQTVPFKK289 pKa = 10.91NVNQIIFPTCAITPATAVCEE309 pKa = 4.07LATTSYY315 pKa = 9.61TASSDD320 pKa = 3.57VADD323 pKa = 3.92ATFVWSITGDD333 pKa = 3.58GTIVDD338 pKa = 4.11AASDD342 pKa = 3.77GTKK345 pKa = 10.14TIVASGGATSTSVSVLAGSAGSYY368 pKa = 8.33TLSVSISKK376 pKa = 8.97TGFGTQTCSQEE387 pKa = 3.56ITVNDD392 pKa = 4.01VPTVTAGADD401 pKa = 3.4QTVCAASPIQTLTATATVSTGATLVWYY428 pKa = 9.33DD429 pKa = 3.61AASGGNTVANPTLSSIGTATYY450 pKa = 9.77YY451 pKa = 10.99AQANLGDD458 pKa = 4.28CSSGRR463 pKa = 11.84VPVVLTINEE472 pKa = 4.54TPTVTAGADD481 pKa = 3.4QTVCAASPIQTLTATATVSTGATLVWYY508 pKa = 9.33DD509 pKa = 3.65AASGGNN515 pKa = 3.39
Molecular weight: 52.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B2H6F0|A0A6B2H6F0_9BACT Metal ABC transporter ATP-binding protein OS=Pontibacter fetidus OX=2700082 GN=GWO68_05890 PE=4 SV=1
MM1 pKa = 7.29CTFATRR7 pKa = 11.84SAGRR11 pKa = 11.84EE12 pKa = 3.54EE13 pKa = 4.32RR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84GRR18 pKa = 11.84SGAQEE23 pKa = 4.05TCLHH27 pKa = 6.8FIKK30 pKa = 10.71LVLRR34 pKa = 11.84LVSTAKK40 pKa = 10.14RR41 pKa = 11.84SKK43 pKa = 10.89NYY45 pKa = 10.22FLTRR49 pKa = 11.84LQKK52 pKa = 10.22QKK54 pKa = 11.02QKK56 pKa = 11.2VYY58 pKa = 10.95LCTPNAAAGRR68 pKa = 11.84AGKK71 pKa = 9.41EE72 pKa = 3.79AEE74 pKa = 4.2TQGFFSGFLSGLTPEE89 pKa = 4.47KK90 pKa = 10.59RR91 pKa = 11.84GG92 pKa = 3.47
MM1 pKa = 7.29CTFATRR7 pKa = 11.84SAGRR11 pKa = 11.84EE12 pKa = 3.54EE13 pKa = 4.32RR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84GRR18 pKa = 11.84SGAQEE23 pKa = 4.05TCLHH27 pKa = 6.8FIKK30 pKa = 10.71LVLRR34 pKa = 11.84LVSTAKK40 pKa = 10.14RR41 pKa = 11.84SKK43 pKa = 10.89NYY45 pKa = 10.22FLTRR49 pKa = 11.84LQKK52 pKa = 10.22QKK54 pKa = 11.02QKK56 pKa = 11.2VYY58 pKa = 10.95LCTPNAAAGRR68 pKa = 11.84AGKK71 pKa = 9.41EE72 pKa = 3.79AEE74 pKa = 4.2TQGFFSGFLSGLTPEE89 pKa = 4.47KK90 pKa = 10.59RR91 pKa = 11.84GG92 pKa = 3.47
Molecular weight: 10.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1154467 |
38 |
3618 |
333.4 |
37.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.101 ± 0.048 | 0.741 ± 0.014 |
5.129 ± 0.034 | 6.269 ± 0.052 |
4.45 ± 0.032 | 6.793 ± 0.041 |
1.96 ± 0.02 | 6.469 ± 0.038 |
6.38 ± 0.051 | 9.927 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.021 | 5.025 ± 0.032 |
4.034 ± 0.027 | 4.134 ± 0.027 |
4.252 ± 0.031 | 5.77 ± 0.035 |
6.263 ± 0.063 | 6.79 ± 0.038 |
1.086 ± 0.014 | 4.077 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
