
Winogradskyella sp. PC-19
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella; unclassified Winogradskyella
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
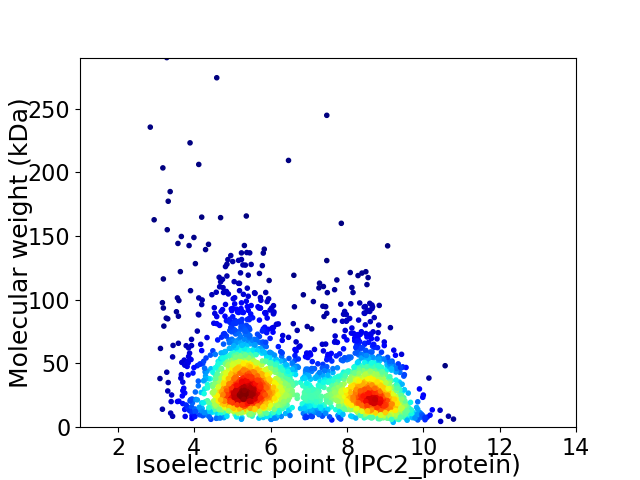
Virtual 2D-PAGE plot for 2717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0MEY8|A0A1Y0MEY8_9FLAO Uncharacterized protein OS=Winogradskyella sp. PC-19 OX=754417 GN=BTO05_02440 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84LYY4 pKa = 11.07LSIIFFLTCSSISFSQLLFDD24 pKa = 4.93DD25 pKa = 4.18EE26 pKa = 4.37AVIRR30 pKa = 11.84GLTGITSYY38 pKa = 11.3DD39 pKa = 3.43GNGNGLSFADD49 pKa = 4.13YY50 pKa = 11.4NNDD53 pKa = 3.29GLDD56 pKa = 4.0DD57 pKa = 3.85VTLPTGNDD65 pKa = 3.16EE66 pKa = 4.13TLKK69 pKa = 10.67FYY71 pKa = 11.42KK72 pKa = 10.21NVNGVFVEE80 pKa = 4.27DD81 pKa = 3.88RR82 pKa = 11.84VLTPAIDD89 pKa = 3.5YY90 pKa = 7.57KK91 pKa = 10.34TRR93 pKa = 11.84SVSWVDD99 pKa = 3.19YY100 pKa = 11.39DD101 pKa = 4.15NDD103 pKa = 3.63GDD105 pKa = 3.88RR106 pKa = 11.84DD107 pKa = 4.0LFVVSDD113 pKa = 3.76IEE115 pKa = 4.25GSRR118 pKa = 11.84LFRR121 pKa = 11.84QDD123 pKa = 3.52PEE125 pKa = 4.33LLFVDD130 pKa = 3.99VTVSSGLFTDD140 pKa = 6.06AIDD143 pKa = 3.89TYY145 pKa = 10.75SVSWGDD151 pKa = 3.66INNDD155 pKa = 2.76GCLDD159 pKa = 3.98LFYY162 pKa = 11.32SNRR165 pKa = 11.84TLNSLITNYY174 pKa = 10.17LFQSNCDD181 pKa = 3.24GTFTNITSTAGLSQDD196 pKa = 3.06PRR198 pKa = 11.84LTFGASFFDD207 pKa = 3.98YY208 pKa = 11.23NNDD211 pKa = 3.26GFQDD215 pKa = 3.38IYY217 pKa = 11.4VINDD221 pKa = 3.27KK222 pKa = 11.1NGSNFLYY229 pKa = 10.77EE230 pKa = 4.14NDD232 pKa = 3.75GDD234 pKa = 4.26STFTDD239 pKa = 3.75VSSQTSTGIVVDD251 pKa = 4.24AMSVTIDD258 pKa = 4.55DD259 pKa = 4.22YY260 pKa = 11.97NSDD263 pKa = 4.17GFFDD267 pKa = 4.0IYY269 pKa = 9.99ITNTPSDD276 pKa = 3.75VATPTLGSVLLKK288 pKa = 10.61NVNGQYY294 pKa = 10.49FEE296 pKa = 5.25DD297 pKa = 4.1VSSSSGTLLDD307 pKa = 3.97GWCWGSNFFDD317 pKa = 5.1ADD319 pKa = 3.21NDD321 pKa = 3.69MDD323 pKa = 4.91LDD325 pKa = 4.67LYY327 pKa = 10.71VSCIYY332 pKa = 9.42TLPGEE337 pKa = 4.18RR338 pKa = 11.84QSYY341 pKa = 9.59GFYY344 pKa = 10.74QNDD347 pKa = 3.53TLEE350 pKa = 4.46VFSEE354 pKa = 4.05PTNIGFANNDD364 pKa = 3.22IDD366 pKa = 3.91SHH368 pKa = 7.57GSVIGDD374 pKa = 3.6ADD376 pKa = 3.77NDD378 pKa = 4.09GKK380 pKa = 11.47VDD382 pKa = 3.31IAVVNNGGLVPNLWMNKK399 pKa = 7.41TVTTNNYY406 pKa = 8.44LTISLEE412 pKa = 4.12GTTSNKK418 pKa = 10.48DD419 pKa = 3.41GIGSKK424 pKa = 9.95IEE426 pKa = 3.63ISINGVKK433 pKa = 9.86QYY435 pKa = 10.8RR436 pKa = 11.84YY437 pKa = 9.47VINGEE442 pKa = 4.58GYY444 pKa = 10.74LSQNSFKK451 pKa = 10.94EE452 pKa = 4.05FFGLGEE458 pKa = 4.21NTVVDD463 pKa = 3.91YY464 pKa = 11.57VKK466 pKa = 10.81VYY468 pKa = 9.17WLSGTIDD475 pKa = 3.59TILNVTANQILNIVEE490 pKa = 4.15GSNVLSNPNLEE501 pKa = 3.85ISEE504 pKa = 4.91DD505 pKa = 4.05FLIHH509 pKa = 6.74PNPTSDD515 pKa = 3.24RR516 pKa = 11.84LSFKK520 pKa = 10.47SRR522 pKa = 11.84NIIEE526 pKa = 4.9TITMYY531 pKa = 10.83NVLGQEE537 pKa = 4.33VFKK540 pKa = 10.89SYY542 pKa = 11.2YY543 pKa = 10.44DD544 pKa = 3.6SLEE547 pKa = 3.68VDD549 pKa = 3.13IDD551 pKa = 3.52ISNLSSGAYY560 pKa = 8.78FVKK563 pKa = 10.1TSLSGVIKK571 pKa = 9.18TSRR574 pKa = 11.84IIKK577 pKa = 9.26QQ578 pKa = 3.04
MM1 pKa = 7.86RR2 pKa = 11.84LYY4 pKa = 11.07LSIIFFLTCSSISFSQLLFDD24 pKa = 4.93DD25 pKa = 4.18EE26 pKa = 4.37AVIRR30 pKa = 11.84GLTGITSYY38 pKa = 11.3DD39 pKa = 3.43GNGNGLSFADD49 pKa = 4.13YY50 pKa = 11.4NNDD53 pKa = 3.29GLDD56 pKa = 4.0DD57 pKa = 3.85VTLPTGNDD65 pKa = 3.16EE66 pKa = 4.13TLKK69 pKa = 10.67FYY71 pKa = 11.42KK72 pKa = 10.21NVNGVFVEE80 pKa = 4.27DD81 pKa = 3.88RR82 pKa = 11.84VLTPAIDD89 pKa = 3.5YY90 pKa = 7.57KK91 pKa = 10.34TRR93 pKa = 11.84SVSWVDD99 pKa = 3.19YY100 pKa = 11.39DD101 pKa = 4.15NDD103 pKa = 3.63GDD105 pKa = 3.88RR106 pKa = 11.84DD107 pKa = 4.0LFVVSDD113 pKa = 3.76IEE115 pKa = 4.25GSRR118 pKa = 11.84LFRR121 pKa = 11.84QDD123 pKa = 3.52PEE125 pKa = 4.33LLFVDD130 pKa = 3.99VTVSSGLFTDD140 pKa = 6.06AIDD143 pKa = 3.89TYY145 pKa = 10.75SVSWGDD151 pKa = 3.66INNDD155 pKa = 2.76GCLDD159 pKa = 3.98LFYY162 pKa = 11.32SNRR165 pKa = 11.84TLNSLITNYY174 pKa = 10.17LFQSNCDD181 pKa = 3.24GTFTNITSTAGLSQDD196 pKa = 3.06PRR198 pKa = 11.84LTFGASFFDD207 pKa = 3.98YY208 pKa = 11.23NNDD211 pKa = 3.26GFQDD215 pKa = 3.38IYY217 pKa = 11.4VINDD221 pKa = 3.27KK222 pKa = 11.1NGSNFLYY229 pKa = 10.77EE230 pKa = 4.14NDD232 pKa = 3.75GDD234 pKa = 4.26STFTDD239 pKa = 3.75VSSQTSTGIVVDD251 pKa = 4.24AMSVTIDD258 pKa = 4.55DD259 pKa = 4.22YY260 pKa = 11.97NSDD263 pKa = 4.17GFFDD267 pKa = 4.0IYY269 pKa = 9.99ITNTPSDD276 pKa = 3.75VATPTLGSVLLKK288 pKa = 10.61NVNGQYY294 pKa = 10.49FEE296 pKa = 5.25DD297 pKa = 4.1VSSSSGTLLDD307 pKa = 3.97GWCWGSNFFDD317 pKa = 5.1ADD319 pKa = 3.21NDD321 pKa = 3.69MDD323 pKa = 4.91LDD325 pKa = 4.67LYY327 pKa = 10.71VSCIYY332 pKa = 9.42TLPGEE337 pKa = 4.18RR338 pKa = 11.84QSYY341 pKa = 9.59GFYY344 pKa = 10.74QNDD347 pKa = 3.53TLEE350 pKa = 4.46VFSEE354 pKa = 4.05PTNIGFANNDD364 pKa = 3.22IDD366 pKa = 3.91SHH368 pKa = 7.57GSVIGDD374 pKa = 3.6ADD376 pKa = 3.77NDD378 pKa = 4.09GKK380 pKa = 11.47VDD382 pKa = 3.31IAVVNNGGLVPNLWMNKK399 pKa = 7.41TVTTNNYY406 pKa = 8.44LTISLEE412 pKa = 4.12GTTSNKK418 pKa = 10.48DD419 pKa = 3.41GIGSKK424 pKa = 9.95IEE426 pKa = 3.63ISINGVKK433 pKa = 9.86QYY435 pKa = 10.8RR436 pKa = 11.84YY437 pKa = 9.47VINGEE442 pKa = 4.58GYY444 pKa = 10.74LSQNSFKK451 pKa = 10.94EE452 pKa = 4.05FFGLGEE458 pKa = 4.21NTVVDD463 pKa = 3.91YY464 pKa = 11.57VKK466 pKa = 10.81VYY468 pKa = 9.17WLSGTIDD475 pKa = 3.59TILNVTANQILNIVEE490 pKa = 4.15GSNVLSNPNLEE501 pKa = 3.85ISEE504 pKa = 4.91DD505 pKa = 4.05FLIHH509 pKa = 6.74PNPTSDD515 pKa = 3.24RR516 pKa = 11.84LSFKK520 pKa = 10.47SRR522 pKa = 11.84NIIEE526 pKa = 4.9TITMYY531 pKa = 10.83NVLGQEE537 pKa = 4.33VFKK540 pKa = 10.89SYY542 pKa = 11.2YY543 pKa = 10.44DD544 pKa = 3.6SLEE547 pKa = 3.68VDD549 pKa = 3.13IDD551 pKa = 3.52ISNLSSGAYY560 pKa = 8.78FVKK563 pKa = 10.1TSLSGVIKK571 pKa = 9.18TSRR574 pKa = 11.84IIKK577 pKa = 9.26QQ578 pKa = 3.04
Molecular weight: 63.96 kDa
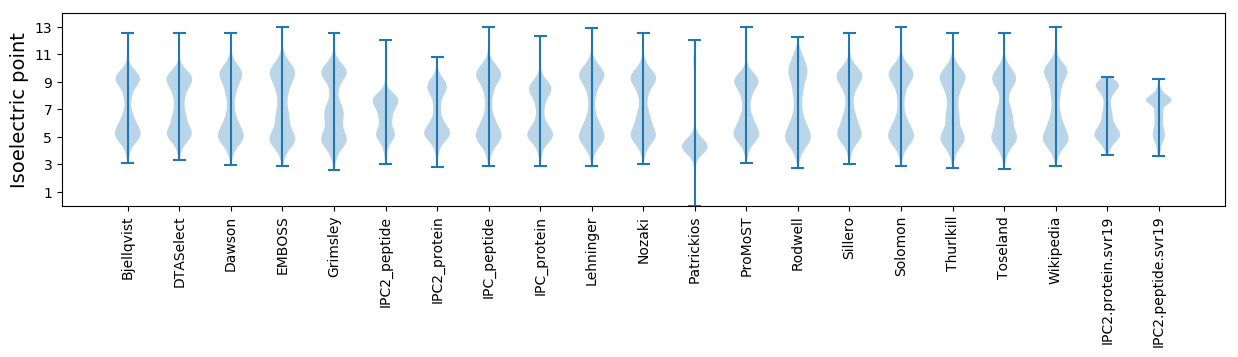
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0MEZ2|A0A1Y0MEZ2_9FLAO MarR family transcriptional regulator OS=Winogradskyella sp. PC-19 OX=754417 GN=BTO05_04310 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 8.18RR42 pKa = 11.84LSVSSDD48 pKa = 3.05LRR50 pKa = 11.84HH51 pKa = 6.32KK52 pKa = 10.54KK53 pKa = 10.01
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 8.18RR42 pKa = 11.84LSVSSDD48 pKa = 3.05LRR50 pKa = 11.84HH51 pKa = 6.32KK52 pKa = 10.54KK53 pKa = 10.01
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910443 |
33 |
2739 |
335.1 |
37.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.166 ± 0.046 | 0.736 ± 0.017 |
5.947 ± 0.042 | 6.445 ± 0.045 |
5.358 ± 0.044 | 6.226 ± 0.046 |
1.614 ± 0.021 | 8.16 ± 0.049 |
7.924 ± 0.075 | 9.142 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.024 | 6.495 ± 0.06 |
3.235 ± 0.029 | 3.27 ± 0.025 |
3.39 ± 0.037 | 6.638 ± 0.041 |
5.906 ± 0.071 | 6.222 ± 0.034 |
0.994 ± 0.014 | 4.045 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
