
Polygala garcinii associated virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; unclassified Geminiviridae
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
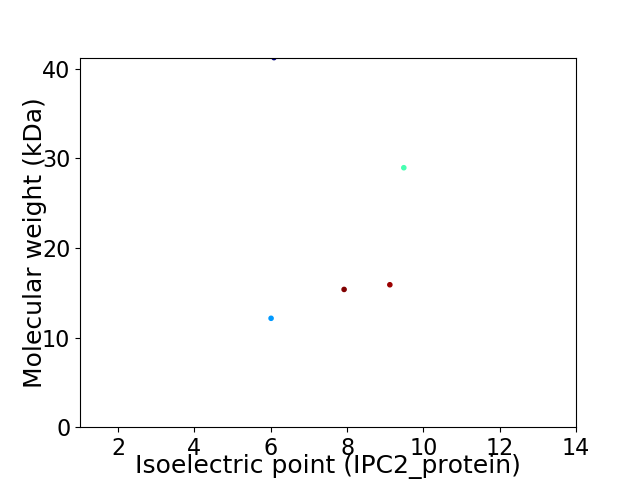
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
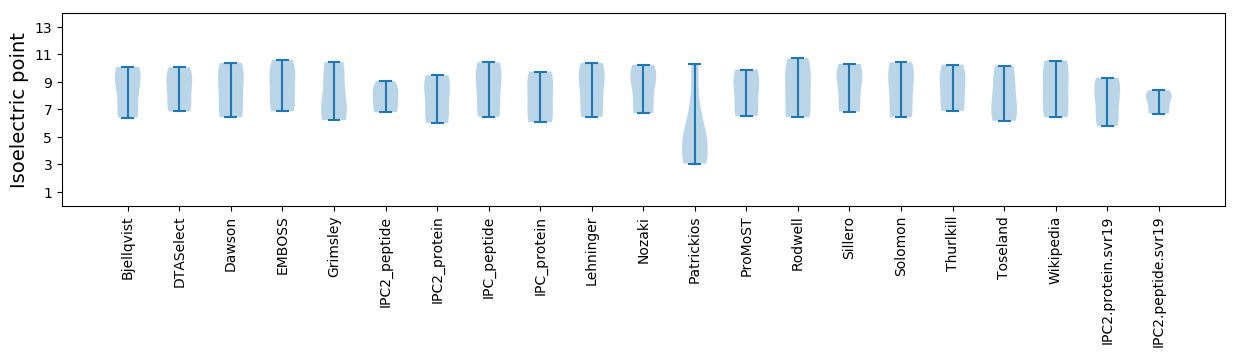
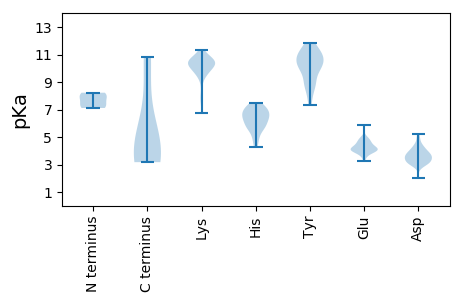
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I8B2Q1|A0A2I8B2Q1_9GEMI Movement protein OS=Polygala garcinii associated virus OX=2093274 PE=3 SV=1
MM1 pKa = 8.08WDD3 pKa = 3.75PLLHH7 pKa = 7.02DD8 pKa = 4.87FPEE11 pKa = 4.45SLHH14 pKa = 6.37GFRR17 pKa = 11.84CMLAVKK23 pKa = 10.28YY24 pKa = 8.24LLCVSSLYY32 pKa = 9.93PVQSVGFEE40 pKa = 4.19LLRR43 pKa = 11.84DD44 pKa = 4.31CISAVRR50 pKa = 11.84TRR52 pKa = 11.84SYY54 pKa = 11.77DD55 pKa = 3.46EE56 pKa = 4.79GSCKK60 pKa = 10.38YY61 pKa = 10.26SALCAKK67 pKa = 9.68VRR69 pKa = 11.84STSEE73 pKa = 3.95GEE75 pKa = 3.98LTQTRR80 pKa = 11.84TPLCCCPHH88 pKa = 6.83CPRR91 pKa = 11.84HH92 pKa = 5.72IGRR95 pKa = 11.84DD96 pKa = 3.31RR97 pKa = 11.84VEE99 pKa = 4.32TEE101 pKa = 3.99TKK103 pKa = 10.4EE104 pKa = 4.12PEE106 pKa = 3.93PP107 pKa = 4.53
MM1 pKa = 8.08WDD3 pKa = 3.75PLLHH7 pKa = 7.02DD8 pKa = 4.87FPEE11 pKa = 4.45SLHH14 pKa = 6.37GFRR17 pKa = 11.84CMLAVKK23 pKa = 10.28YY24 pKa = 8.24LLCVSSLYY32 pKa = 9.93PVQSVGFEE40 pKa = 4.19LLRR43 pKa = 11.84DD44 pKa = 4.31CISAVRR50 pKa = 11.84TRR52 pKa = 11.84SYY54 pKa = 11.77DD55 pKa = 3.46EE56 pKa = 4.79GSCKK60 pKa = 10.38YY61 pKa = 10.26SALCAKK67 pKa = 9.68VRR69 pKa = 11.84STSEE73 pKa = 3.95GEE75 pKa = 3.98LTQTRR80 pKa = 11.84TPLCCCPHH88 pKa = 6.83CPRR91 pKa = 11.84HH92 pKa = 5.72IGRR95 pKa = 11.84DD96 pKa = 3.31RR97 pKa = 11.84VEE99 pKa = 4.32TEE101 pKa = 3.99TKK103 pKa = 10.4EE104 pKa = 4.12PEE106 pKa = 3.93PP107 pKa = 4.53
Molecular weight: 12.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I8B2M1|A0A2I8B2M1_9GEMI Replication-associated protein OS=Polygala garcinii associated virus OX=2093274 PE=3 SV=1
MM1 pKa = 7.11TKK3 pKa = 10.3GRR5 pKa = 11.84VSIVPFVRR13 pKa = 11.84RR14 pKa = 11.84LGPRR18 pKa = 11.84LKK20 pKa = 10.82GNSPRR25 pKa = 11.84RR26 pKa = 11.84GPRR29 pKa = 11.84YY30 pKa = 9.24VVARR34 pKa = 11.84TAPVISAVTEE44 pKa = 4.09WKK46 pKa = 10.11RR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84NRR52 pKa = 11.84NPKK55 pKa = 9.42SSKK58 pKa = 10.16KK59 pKa = 10.17DD60 pKa = 3.35VVPFGCEE67 pKa = 4.16GPCKK71 pKa = 10.47VEE73 pKa = 4.28SFSDD77 pKa = 3.52RR78 pKa = 11.84YY79 pKa = 8.86EE80 pKa = 4.11VKK82 pKa = 9.2HH83 pKa = 6.07TGTVCCLSQISRR95 pKa = 11.84GNGLKK100 pKa = 10.03QRR102 pKa = 11.84IGKK105 pKa = 9.95RR106 pKa = 11.84ITIKK110 pKa = 10.5SIYY113 pKa = 10.08LLGKK117 pKa = 9.56VWLDD121 pKa = 3.05GSNRR125 pKa = 11.84ISNHH129 pKa = 4.4TNVCIFFVVRR139 pKa = 11.84DD140 pKa = 3.97RR141 pKa = 11.84RR142 pKa = 11.84PSVTPINFGSLFDD155 pKa = 4.09MFDD158 pKa = 3.88GEE160 pKa = 4.39PATASVKK167 pKa = 10.18SDD169 pKa = 2.8HH170 pKa = 7.13RR171 pKa = 11.84DD172 pKa = 3.17RR173 pKa = 11.84FQVLYY178 pKa = 9.82RR179 pKa = 11.84FQVSVSGGQYY189 pKa = 9.94SSRR192 pKa = 11.84DD193 pKa = 3.07QTVFQKK199 pKa = 10.29FVNLNHH205 pKa = 6.56RR206 pKa = 11.84VVFDD210 pKa = 3.7NQDD213 pKa = 3.2DD214 pKa = 4.4GVYY217 pKa = 10.91GNHH220 pKa = 6.87DD221 pKa = 3.58QNALLVYY228 pKa = 7.32MACCHH233 pKa = 4.93STNWLYY239 pKa = 11.66SSMKK243 pKa = 10.04AKK245 pKa = 10.62VYY247 pKa = 10.2FYY249 pKa = 10.97DD250 pKa = 3.62SAMNN254 pKa = 3.79
MM1 pKa = 7.11TKK3 pKa = 10.3GRR5 pKa = 11.84VSIVPFVRR13 pKa = 11.84RR14 pKa = 11.84LGPRR18 pKa = 11.84LKK20 pKa = 10.82GNSPRR25 pKa = 11.84RR26 pKa = 11.84GPRR29 pKa = 11.84YY30 pKa = 9.24VVARR34 pKa = 11.84TAPVISAVTEE44 pKa = 4.09WKK46 pKa = 10.11RR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84NRR52 pKa = 11.84NPKK55 pKa = 9.42SSKK58 pKa = 10.16KK59 pKa = 10.17DD60 pKa = 3.35VVPFGCEE67 pKa = 4.16GPCKK71 pKa = 10.47VEE73 pKa = 4.28SFSDD77 pKa = 3.52RR78 pKa = 11.84YY79 pKa = 8.86EE80 pKa = 4.11VKK82 pKa = 9.2HH83 pKa = 6.07TGTVCCLSQISRR95 pKa = 11.84GNGLKK100 pKa = 10.03QRR102 pKa = 11.84IGKK105 pKa = 9.95RR106 pKa = 11.84ITIKK110 pKa = 10.5SIYY113 pKa = 10.08LLGKK117 pKa = 9.56VWLDD121 pKa = 3.05GSNRR125 pKa = 11.84ISNHH129 pKa = 4.4TNVCIFFVVRR139 pKa = 11.84DD140 pKa = 3.97RR141 pKa = 11.84RR142 pKa = 11.84PSVTPINFGSLFDD155 pKa = 4.09MFDD158 pKa = 3.88GEE160 pKa = 4.39PATASVKK167 pKa = 10.18SDD169 pKa = 2.8HH170 pKa = 7.13RR171 pKa = 11.84DD172 pKa = 3.17RR173 pKa = 11.84FQVLYY178 pKa = 9.82RR179 pKa = 11.84FQVSVSGGQYY189 pKa = 9.94SSRR192 pKa = 11.84DD193 pKa = 3.07QTVFQKK199 pKa = 10.29FVNLNHH205 pKa = 6.56RR206 pKa = 11.84VVFDD210 pKa = 3.7NQDD213 pKa = 3.2DD214 pKa = 4.4GVYY217 pKa = 10.91GNHH220 pKa = 6.87DD221 pKa = 3.58QNALLVYY228 pKa = 7.32MACCHH233 pKa = 4.93STNWLYY239 pKa = 11.66SSMKK243 pKa = 10.04AKK245 pKa = 10.62VYY247 pKa = 10.2FYY249 pKa = 10.97DD250 pKa = 3.62SAMNN254 pKa = 3.79
Molecular weight: 28.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992 |
107 |
362 |
198.4 |
22.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.435 ± 0.334 | 3.125 ± 0.763 |
5.746 ± 0.246 | 4.536 ± 1.155 |
4.839 ± 0.373 | 5.444 ± 0.635 |
4.133 ± 0.871 | 4.738 ± 0.497 |
5.645 ± 0.536 | 7.258 ± 1.276 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.512 ± 0.165 | 4.839 ± 0.782 |
6.048 ± 0.697 | 4.234 ± 0.528 |
7.359 ± 0.941 | 7.56 ± 0.712 |
6.754 ± 0.723 | 6.754 ± 1.32 |
1.21 ± 0.095 | 3.831 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
