
Paraburkholderia megapolitana
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
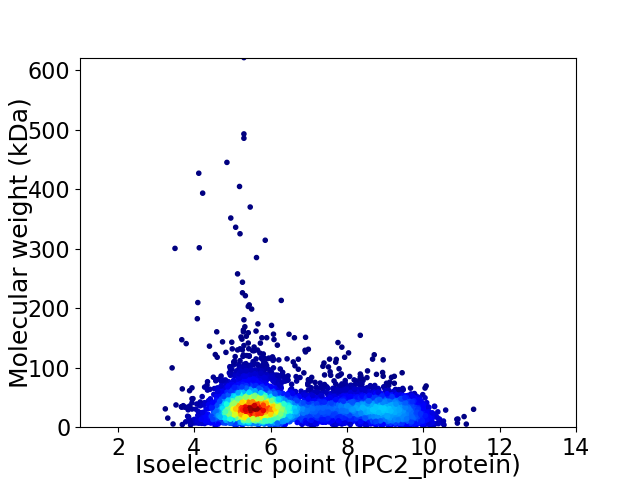
Virtual 2D-PAGE plot for 6692 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
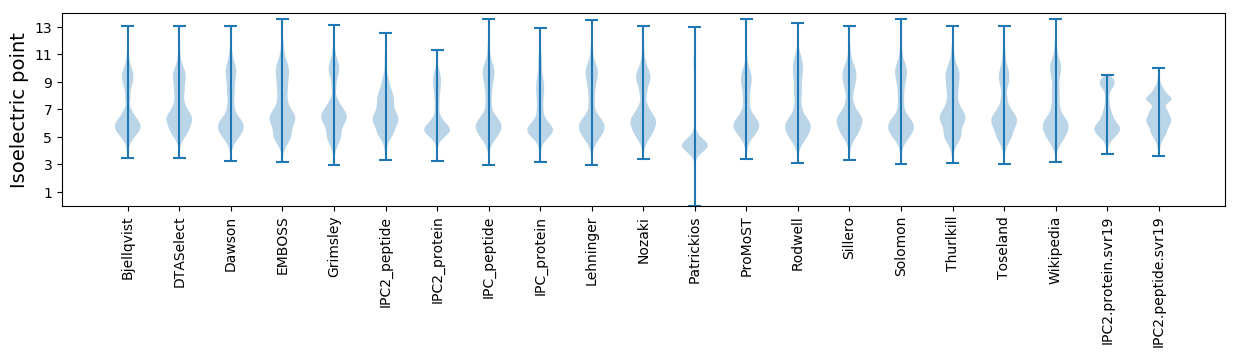
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3DVB0|A0A1I3DVB0_9BURK Type IV pilus biogenesis protein PilP OS=Paraburkholderia megapolitana OX=420953 GN=SAMN05192543_101544 PE=4 SV=1
MM1 pKa = 7.49SSAAVQGYY9 pKa = 7.17STNPMDD15 pKa = 4.22YY16 pKa = 10.67LSNSNSYY23 pKa = 11.06GYY25 pKa = 10.71GSSDD29 pKa = 2.79ISGEE33 pKa = 4.13SGGVNGLNGNNSGTGMNEE51 pKa = 3.63QQILQIIEE59 pKa = 4.6EE60 pKa = 4.59LLQMLQQVLQNSDD73 pKa = 3.4GDD75 pKa = 4.13DD76 pKa = 3.77SGGGAPPVGSGSSPGGSSNPFPQDD100 pKa = 3.18YY101 pKa = 10.94SSAPPVGSGGNAPPLANSQSPATNGAPPVSGNAPPVTSNAPPPPSNPPPVNSGGNNSGNNGAPPAGSSTQTTNNPPPPANGSSGSSSGPTMFGLSTAPQTTALSQSQGQQVADD214 pKa = 3.34QYY216 pKa = 11.87VNNLMSDD223 pKa = 4.32FGLTKK228 pKa = 10.1PQAQGIVANLWHH240 pKa = 7.01EE241 pKa = 4.35SGGMNSGINQGGQIGAPTGNNADD264 pKa = 4.49DD265 pKa = 3.76NANGYY270 pKa = 10.39GIAQWGGSRR279 pKa = 11.84KK280 pKa = 9.74EE281 pKa = 3.93GLEE284 pKa = 4.1AYY286 pKa = 9.73AKK288 pKa = 10.65QNGLDD293 pKa = 3.87PSSEE297 pKa = 3.88AANYY301 pKa = 10.17GYY303 pKa = 10.68LKK305 pKa = 10.51QEE307 pKa = 4.15LQTTQSGAISAVKK320 pKa = 9.01GTSNAQDD327 pKa = 3.16ATSAFMTSFEE337 pKa = 4.54KK338 pKa = 10.76PSDD341 pKa = 3.68PEE343 pKa = 4.02LASRR347 pKa = 11.84LADD350 pKa = 3.62LSLVQGG356 pKa = 3.98
MM1 pKa = 7.49SSAAVQGYY9 pKa = 7.17STNPMDD15 pKa = 4.22YY16 pKa = 10.67LSNSNSYY23 pKa = 11.06GYY25 pKa = 10.71GSSDD29 pKa = 2.79ISGEE33 pKa = 4.13SGGVNGLNGNNSGTGMNEE51 pKa = 3.63QQILQIIEE59 pKa = 4.6EE60 pKa = 4.59LLQMLQQVLQNSDD73 pKa = 3.4GDD75 pKa = 4.13DD76 pKa = 3.77SGGGAPPVGSGSSPGGSSNPFPQDD100 pKa = 3.18YY101 pKa = 10.94SSAPPVGSGGNAPPLANSQSPATNGAPPVSGNAPPVTSNAPPPPSNPPPVNSGGNNSGNNGAPPAGSSTQTTNNPPPPANGSSGSSSGPTMFGLSTAPQTTALSQSQGQQVADD214 pKa = 3.34QYY216 pKa = 11.87VNNLMSDD223 pKa = 4.32FGLTKK228 pKa = 10.1PQAQGIVANLWHH240 pKa = 7.01EE241 pKa = 4.35SGGMNSGINQGGQIGAPTGNNADD264 pKa = 4.49DD265 pKa = 3.76NANGYY270 pKa = 10.39GIAQWGGSRR279 pKa = 11.84KK280 pKa = 9.74EE281 pKa = 3.93GLEE284 pKa = 4.1AYY286 pKa = 9.73AKK288 pKa = 10.65QNGLDD293 pKa = 3.87PSSEE297 pKa = 3.88AANYY301 pKa = 10.17GYY303 pKa = 10.68LKK305 pKa = 10.51QEE307 pKa = 4.15LQTTQSGAISAVKK320 pKa = 9.01GTSNAQDD327 pKa = 3.16ATSAFMTSFEE337 pKa = 4.54KK338 pKa = 10.76PSDD341 pKa = 3.68PEE343 pKa = 4.02LASRR347 pKa = 11.84LADD350 pKa = 3.62LSLVQGG356 pKa = 3.98
Molecular weight: 35.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3G0T1|A0A1I3G0T1_9BURK Protoheme IX farnesyltransferase OS=Paraburkholderia megapolitana OX=420953 GN=ctaB PE=3 SV=1
MM1 pKa = 7.24NRR3 pKa = 11.84QFKK6 pKa = 7.92MTTIAMCVSSLIALSGCGSTVTKK29 pKa = 10.2PLGSGSGGGTIGTGGGGGGTGTGGGGTGTGGGGGKK64 pKa = 8.0TPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTLRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84PLQLLPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84PRR141 pKa = 11.84LLRR144 pKa = 11.84RR145 pKa = 11.84LLRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84HH151 pKa = 5.26RR152 pKa = 11.84RR153 pKa = 11.84LLRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84LLRR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84LRR166 pKa = 11.84RR167 pKa = 11.84LLRR170 pKa = 11.84RR171 pKa = 11.84PLRR174 pKa = 11.84RR175 pKa = 11.84LRR177 pKa = 11.84LRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LTPTPSATSSRR192 pKa = 11.84TPATSSPRR200 pKa = 11.84LARR203 pKa = 11.84LSPRR207 pKa = 11.84SAARR211 pKa = 11.84SVSRR215 pKa = 11.84RR216 pKa = 11.84FPAATPRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84LRR227 pKa = 11.84SATSSPTSAPVSLRR241 pKa = 11.84SAMEE245 pKa = 3.99RR246 pKa = 11.84PTVSASSARR255 pKa = 11.84LLTRR259 pKa = 11.84SASPSPAQAMSSTTPARR276 pKa = 11.84PSTTLGSS283 pKa = 3.55
MM1 pKa = 7.24NRR3 pKa = 11.84QFKK6 pKa = 7.92MTTIAMCVSSLIALSGCGSTVTKK29 pKa = 10.2PLGSGSGGGTIGTGGGGGGTGTGGGGTGTGGGGGKK64 pKa = 8.0TPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTLRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84PLQLLPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84PRR141 pKa = 11.84LLRR144 pKa = 11.84RR145 pKa = 11.84LLRR148 pKa = 11.84RR149 pKa = 11.84RR150 pKa = 11.84HH151 pKa = 5.26RR152 pKa = 11.84RR153 pKa = 11.84LLRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84LLRR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84LRR166 pKa = 11.84RR167 pKa = 11.84LLRR170 pKa = 11.84RR171 pKa = 11.84PLRR174 pKa = 11.84RR175 pKa = 11.84LRR177 pKa = 11.84LRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LTPTPSATSSRR192 pKa = 11.84TPATSSPRR200 pKa = 11.84LARR203 pKa = 11.84LSPRR207 pKa = 11.84SAARR211 pKa = 11.84SVSRR215 pKa = 11.84RR216 pKa = 11.84FPAATPRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84LRR227 pKa = 11.84SATSSPTSAPVSLRR241 pKa = 11.84SAMEE245 pKa = 3.99RR246 pKa = 11.84PTVSASSARR255 pKa = 11.84LLTRR259 pKa = 11.84SASPSPAQAMSSTTPARR276 pKa = 11.84PSTTLGSS283 pKa = 3.55
Molecular weight: 30.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2228779 |
25 |
5663 |
333.1 |
36.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.541 ± 0.044 | 0.89 ± 0.009 |
5.504 ± 0.026 | 4.854 ± 0.033 |
3.686 ± 0.018 | 8.229 ± 0.045 |
2.351 ± 0.015 | 4.732 ± 0.024 |
2.826 ± 0.025 | 10.321 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.014 | 2.909 ± 0.027 |
5.138 ± 0.024 | 3.694 ± 0.02 |
6.723 ± 0.036 | 5.895 ± 0.029 |
5.836 ± 0.037 | 7.782 ± 0.029 |
1.405 ± 0.011 | 2.471 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
