
Caenorhabditis briggsae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Peloderinae; Caenorhabditis
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
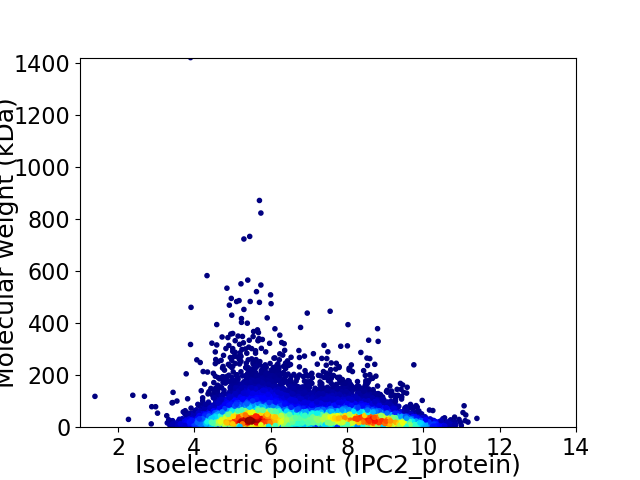
Virtual 2D-PAGE plot for 21755 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
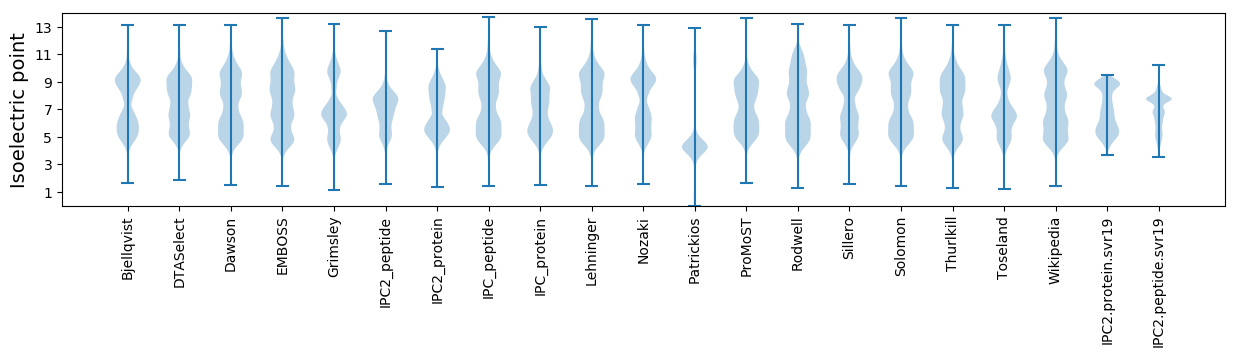
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2J6X4|G2J6X4_CAEBR Protein CBG17140 OS=Caenorhabditis briggsae OX=6238 GN=CBG17140 PE=4 SV=1
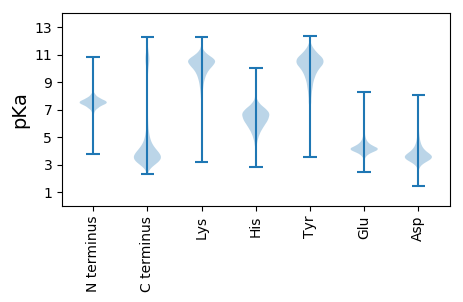
MM1 pKa = 7.54PRR3 pKa = 11.84YY4 pKa = 9.99SLLLLIIIPIANGYY18 pKa = 9.46IFDD21 pKa = 4.27PSRR24 pKa = 11.84TRR26 pKa = 11.84KK27 pKa = 9.8VLIVPKK33 pKa = 10.46NKK35 pKa = 10.4NGWHH39 pKa = 7.16DD40 pKa = 3.75SATFAPIDD48 pKa = 4.5PIQIGTQDD56 pKa = 3.41WEE58 pKa = 4.44DD59 pKa = 3.42LTEE62 pKa = 3.92VEE64 pKa = 5.03RR65 pKa = 11.84YY66 pKa = 9.41YY67 pKa = 11.37LEE69 pKa = 4.14TKK71 pKa = 9.02GTLRR75 pKa = 11.84FSGSSAYY82 pKa = 10.27DD83 pKa = 3.24YY84 pKa = 11.67GNQEE88 pKa = 4.46LKK90 pKa = 10.79DD91 pKa = 4.42DD92 pKa = 4.54YY93 pKa = 11.55DD94 pKa = 4.06VSDD97 pKa = 5.15YY98 pKa = 11.02IDD100 pKa = 4.54EE101 pKa = 4.32EE102 pKa = 4.41DD103 pKa = 5.88KK104 pKa = 11.51EE105 pKa = 5.23DD106 pKa = 4.86DD107 pKa = 4.06DD108 pKa = 4.91EE109 pKa = 6.35KK110 pKa = 11.55GSGDD114 pKa = 3.54YY115 pKa = 11.47GDD117 pKa = 3.82VSKK120 pKa = 11.15EE121 pKa = 4.18DD122 pKa = 3.6EE123 pKa = 4.24DD124 pKa = 4.55SEE126 pKa = 5.0YY127 pKa = 11.01EE128 pKa = 4.36GYY130 pKa = 10.9LEE132 pKa = 6.29DD133 pKa = 5.82EE134 pKa = 4.69FDD136 pKa = 5.63DD137 pKa = 4.07YY138 pKa = 12.06DD139 pKa = 3.86NQYY142 pKa = 11.24EE143 pKa = 4.19YY144 pKa = 11.48DD145 pKa = 3.59NYY147 pKa = 11.67
MM1 pKa = 7.54PRR3 pKa = 11.84YY4 pKa = 9.99SLLLLIIIPIANGYY18 pKa = 9.46IFDD21 pKa = 4.27PSRR24 pKa = 11.84TRR26 pKa = 11.84KK27 pKa = 9.8VLIVPKK33 pKa = 10.46NKK35 pKa = 10.4NGWHH39 pKa = 7.16DD40 pKa = 3.75SATFAPIDD48 pKa = 4.5PIQIGTQDD56 pKa = 3.41WEE58 pKa = 4.44DD59 pKa = 3.42LTEE62 pKa = 3.92VEE64 pKa = 5.03RR65 pKa = 11.84YY66 pKa = 9.41YY67 pKa = 11.37LEE69 pKa = 4.14TKK71 pKa = 9.02GTLRR75 pKa = 11.84FSGSSAYY82 pKa = 10.27DD83 pKa = 3.24YY84 pKa = 11.67GNQEE88 pKa = 4.46LKK90 pKa = 10.79DD91 pKa = 4.42DD92 pKa = 4.54YY93 pKa = 11.55DD94 pKa = 4.06VSDD97 pKa = 5.15YY98 pKa = 11.02IDD100 pKa = 4.54EE101 pKa = 4.32EE102 pKa = 4.41DD103 pKa = 5.88KK104 pKa = 11.51EE105 pKa = 5.23DD106 pKa = 4.86DD107 pKa = 4.06DD108 pKa = 4.91EE109 pKa = 6.35KK110 pKa = 11.55GSGDD114 pKa = 3.54YY115 pKa = 11.47GDD117 pKa = 3.82VSKK120 pKa = 11.15EE121 pKa = 4.18DD122 pKa = 3.6EE123 pKa = 4.24DD124 pKa = 4.55SEE126 pKa = 5.0YY127 pKa = 11.01EE128 pKa = 4.36GYY130 pKa = 10.9LEE132 pKa = 6.29DD133 pKa = 5.82EE134 pKa = 4.69FDD136 pKa = 5.63DD137 pKa = 4.07YY138 pKa = 12.06DD139 pKa = 3.86NQYY142 pKa = 11.24EE143 pKa = 4.19YY144 pKa = 11.48DD145 pKa = 3.59NYY147 pKa = 11.67
Molecular weight: 17.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8Y3V3|A8Y3V3_CAEBR Protein CBR-SNB-2 OS=Caenorhabditis briggsae OX=6238 GN=snb-2 PE=4 SV=1
LL1 pKa = 6.68ITPSPAPIPALPVPIPHH18 pKa = 7.37RR19 pKa = 11.84PTPIPPPSALIQSPPAPIPPSPASIPPSPAPIPHH53 pKa = 7.35PLAPIPLTPAAIPLPSAPIPPPPAPIQSSPALIPPPPAPIPPPPALIPPPPALIPPPPAPIPPSPAPIPPSPAPIPPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPSSPIPPPPAPIPPPPAPIPPPILRR198 pKa = 11.84FHH200 pKa = 6.61LHH202 pKa = 4.54QLRR205 pKa = 11.84FHH207 pKa = 6.51LHH209 pKa = 4.2QLRR212 pKa = 11.84FHH214 pKa = 6.51LHH216 pKa = 4.2QLRR219 pKa = 11.84FHH221 pKa = 6.51LHH223 pKa = 4.2QLRR226 pKa = 11.84FHH228 pKa = 6.51LHH230 pKa = 4.2QLRR233 pKa = 11.84FHH235 pKa = 6.51LHH237 pKa = 4.2QLRR240 pKa = 11.84FHH242 pKa = 6.72LHH244 pKa = 5.02QLHH247 pKa = 6.23LHH249 pKa = 4.91QLRR252 pKa = 11.84FHH254 pKa = 6.51LHH256 pKa = 4.2QLRR259 pKa = 11.84FHH261 pKa = 6.51LHH263 pKa = 4.2QLRR266 pKa = 11.84FHH268 pKa = 6.42LHH270 pKa = 5.85HH271 pKa = 6.88LRR273 pKa = 11.84FHH275 pKa = 6.37LHH277 pKa = 4.47QLRR280 pKa = 11.84FHH282 pKa = 6.51LHH284 pKa = 4.2QLRR287 pKa = 11.84FHH289 pKa = 6.51LHH291 pKa = 4.2QLRR294 pKa = 11.84FHH296 pKa = 6.51LHH298 pKa = 4.2QLRR301 pKa = 11.84FHH303 pKa = 6.74LHH305 pKa = 4.42QLL307 pKa = 3.48
LL1 pKa = 6.68ITPSPAPIPALPVPIPHH18 pKa = 7.37RR19 pKa = 11.84PTPIPPPSALIQSPPAPIPPSPASIPPSPAPIPHH53 pKa = 7.35PLAPIPLTPAAIPLPSAPIPPPPAPIQSSPALIPPPPAPIPPPPALIPPPPALIPPPPAPIPPSPAPIPPSPAPIPPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPPAPIPPPSSPIPPPPAPIPPPPAPIPPPILRR198 pKa = 11.84FHH200 pKa = 6.61LHH202 pKa = 4.54QLRR205 pKa = 11.84FHH207 pKa = 6.51LHH209 pKa = 4.2QLRR212 pKa = 11.84FHH214 pKa = 6.51LHH216 pKa = 4.2QLRR219 pKa = 11.84FHH221 pKa = 6.51LHH223 pKa = 4.2QLRR226 pKa = 11.84FHH228 pKa = 6.51LHH230 pKa = 4.2QLRR233 pKa = 11.84FHH235 pKa = 6.51LHH237 pKa = 4.2QLRR240 pKa = 11.84FHH242 pKa = 6.72LHH244 pKa = 5.02QLHH247 pKa = 6.23LHH249 pKa = 4.91QLRR252 pKa = 11.84FHH254 pKa = 6.51LHH256 pKa = 4.2QLRR259 pKa = 11.84FHH261 pKa = 6.51LHH263 pKa = 4.2QLRR266 pKa = 11.84FHH268 pKa = 6.42LHH270 pKa = 5.85HH271 pKa = 6.88LRR273 pKa = 11.84FHH275 pKa = 6.37LHH277 pKa = 4.47QLRR280 pKa = 11.84FHH282 pKa = 6.51LHH284 pKa = 4.2QLRR287 pKa = 11.84FHH289 pKa = 6.51LHH291 pKa = 4.2QLRR294 pKa = 11.84FHH296 pKa = 6.51LHH298 pKa = 4.2QLRR301 pKa = 11.84FHH303 pKa = 6.74LHH305 pKa = 4.42QLL307 pKa = 3.48
Molecular weight: 33.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8683488 |
8 |
13580 |
399.1 |
45.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.222 ± 0.02 | 1.963 ± 0.015 |
5.288 ± 0.017 | 6.851 ± 0.026 |
4.681 ± 0.016 | 5.342 ± 0.027 |
2.276 ± 0.009 | 5.971 ± 0.016 |
6.432 ± 0.02 | 8.476 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.009 | 4.827 ± 0.012 |
5.08 ± 0.025 | 4.16 ± 0.019 |
5.368 ± 0.018 | 8.066 ± 0.025 |
5.788 ± 0.024 | 6.134 ± 0.015 |
1.121 ± 0.006 | 3.118 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
