
Hubei sobemo-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
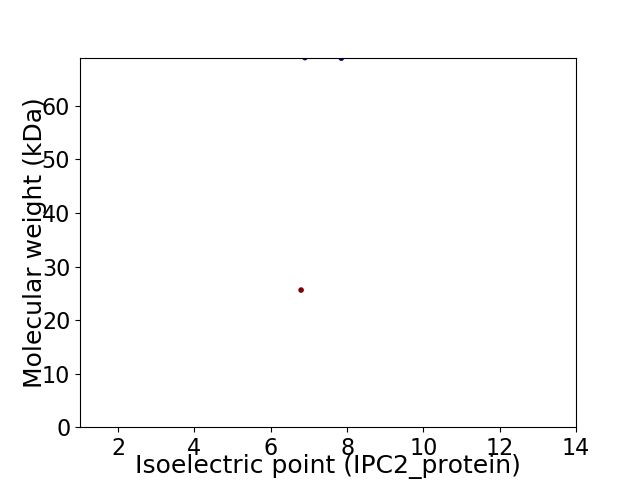
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEM4|A0A1L3KEM4_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 6 OX=1923239 PE=4 SV=1
MM1 pKa = 7.83PDD3 pKa = 3.2WVVEE7 pKa = 4.34AYY9 pKa = 10.16FRR11 pKa = 11.84ARR13 pKa = 11.84LAQCSNSDD21 pKa = 3.09QRR23 pKa = 11.84YY24 pKa = 8.55EE25 pKa = 3.97KK26 pKa = 10.86LCWNRR31 pKa = 11.84MMCVVGPGAVLRR43 pKa = 11.84LPDD46 pKa = 3.96GQRR49 pKa = 11.84LRR51 pKa = 11.84QTCVGFMKK59 pKa = 10.58SGWLLTLSMNSAAQFFQHH77 pKa = 4.48VVAWRR82 pKa = 11.84RR83 pKa = 11.84SGYY86 pKa = 10.25DD87 pKa = 3.62LSSLPYY93 pKa = 8.03MWAMGDD99 pKa = 3.73DD100 pKa = 3.43MLLRR104 pKa = 11.84WEE106 pKa = 4.54EE107 pKa = 4.58GYY109 pKa = 11.09KK110 pKa = 10.37LEE112 pKa = 5.24PYY114 pKa = 10.47VAMLATTGCIVKK126 pKa = 9.79HH127 pKa = 5.47AVWSRR132 pKa = 11.84EE133 pKa = 3.54FAGYY137 pKa = 10.24AFKK140 pKa = 11.16GDD142 pKa = 4.0GVSPLYY148 pKa = 9.4PAKK151 pKa = 10.64HH152 pKa = 6.31KK153 pKa = 9.02FTLSYY158 pKa = 10.64VKK160 pKa = 10.73EE161 pKa = 4.11EE162 pKa = 4.26VEE164 pKa = 4.39QEE166 pKa = 4.09LLLSYY171 pKa = 10.53HH172 pKa = 7.1LLYY175 pKa = 11.13ALDD178 pKa = 4.32PNSWLPAVRR187 pKa = 11.84EE188 pKa = 4.08HH189 pKa = 7.03CRR191 pKa = 11.84FPVGRR196 pKa = 11.84MFRR199 pKa = 11.84MWAMGLIEE207 pKa = 6.14LEE209 pKa = 4.35MLPLMTRR216 pKa = 11.84VGCDD220 pKa = 2.96EE221 pKa = 3.99
MM1 pKa = 7.83PDD3 pKa = 3.2WVVEE7 pKa = 4.34AYY9 pKa = 10.16FRR11 pKa = 11.84ARR13 pKa = 11.84LAQCSNSDD21 pKa = 3.09QRR23 pKa = 11.84YY24 pKa = 8.55EE25 pKa = 3.97KK26 pKa = 10.86LCWNRR31 pKa = 11.84MMCVVGPGAVLRR43 pKa = 11.84LPDD46 pKa = 3.96GQRR49 pKa = 11.84LRR51 pKa = 11.84QTCVGFMKK59 pKa = 10.58SGWLLTLSMNSAAQFFQHH77 pKa = 4.48VVAWRR82 pKa = 11.84RR83 pKa = 11.84SGYY86 pKa = 10.25DD87 pKa = 3.62LSSLPYY93 pKa = 8.03MWAMGDD99 pKa = 3.73DD100 pKa = 3.43MLLRR104 pKa = 11.84WEE106 pKa = 4.54EE107 pKa = 4.58GYY109 pKa = 11.09KK110 pKa = 10.37LEE112 pKa = 5.24PYY114 pKa = 10.47VAMLATTGCIVKK126 pKa = 9.79HH127 pKa = 5.47AVWSRR132 pKa = 11.84EE133 pKa = 3.54FAGYY137 pKa = 10.24AFKK140 pKa = 11.16GDD142 pKa = 4.0GVSPLYY148 pKa = 9.4PAKK151 pKa = 10.64HH152 pKa = 6.31KK153 pKa = 9.02FTLSYY158 pKa = 10.64VKK160 pKa = 10.73EE161 pKa = 4.11EE162 pKa = 4.26VEE164 pKa = 4.39QEE166 pKa = 4.09LLLSYY171 pKa = 10.53HH172 pKa = 7.1LLYY175 pKa = 11.13ALDD178 pKa = 4.32PNSWLPAVRR187 pKa = 11.84EE188 pKa = 4.08HH189 pKa = 7.03CRR191 pKa = 11.84FPVGRR196 pKa = 11.84MFRR199 pKa = 11.84MWAMGLIEE207 pKa = 6.14LEE209 pKa = 4.35MLPLMTRR216 pKa = 11.84VGCDD220 pKa = 2.96EE221 pKa = 3.99
Molecular weight: 25.62 kDa
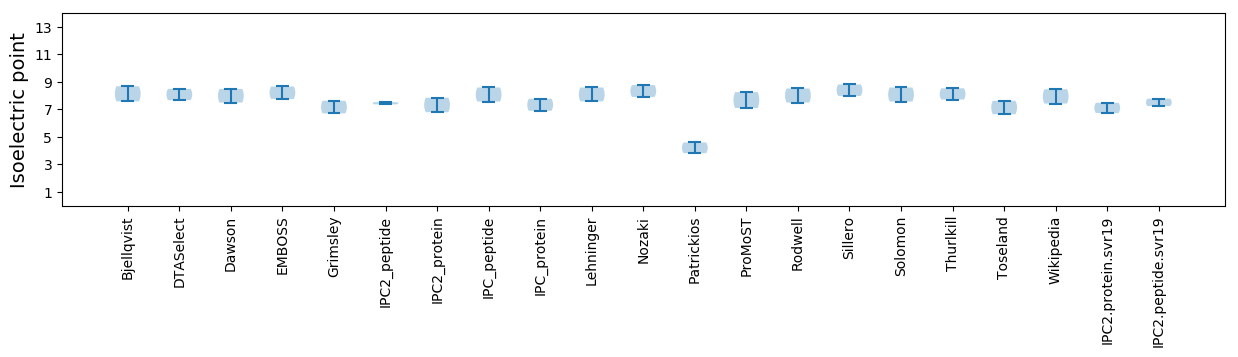
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEM4|A0A1L3KEM4_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 6 OX=1923239 PE=4 SV=1
MM1 pKa = 7.08YY2 pKa = 10.2AVSMYY7 pKa = 10.86LATLLVWAVFMTYY20 pKa = 8.78TVVDD24 pKa = 5.21FFWDD28 pKa = 3.52WPVVRR33 pKa = 11.84QAVLRR38 pKa = 11.84DD39 pKa = 3.51RR40 pKa = 11.84AGKK43 pKa = 10.29ALFDD47 pKa = 3.86FCLGAVVLGIGFVLFFSSHH66 pKa = 7.82DD67 pKa = 3.24ILEE70 pKa = 4.62WIGRR74 pKa = 11.84GVEE77 pKa = 4.06TLVRR81 pKa = 11.84RR82 pKa = 11.84IWKK85 pKa = 9.4FCFARR90 pKa = 11.84ATARR94 pKa = 11.84VARR97 pKa = 11.84PDD99 pKa = 2.92KK100 pKa = 10.95SMRR103 pKa = 11.84RR104 pKa = 11.84VLEE107 pKa = 4.06TMKK110 pKa = 10.6EE111 pKa = 4.17GSDD114 pKa = 3.51LQTIPRR120 pKa = 11.84PACQLALYY128 pKa = 9.8DD129 pKa = 3.74YY130 pKa = 11.35DD131 pKa = 4.32SIEE134 pKa = 3.91RR135 pKa = 11.84QKK137 pKa = 11.25YY138 pKa = 7.83YY139 pKa = 10.73LGGAIHH145 pKa = 6.77VEE147 pKa = 4.24GGWLVSCTHH156 pKa = 6.7IVNQLRR162 pKa = 11.84NKK164 pKa = 9.76PYY166 pKa = 10.94ASVLDD171 pKa = 4.09PQGVEE176 pKa = 3.52QFYY179 pKa = 11.02SLEE182 pKa = 3.95GLEE185 pKa = 4.15WEE187 pKa = 4.86EE188 pKa = 3.84IGADD192 pKa = 3.45VSVAKK197 pKa = 10.55LGRR200 pKa = 11.84VFQHH204 pKa = 6.62LKK206 pKa = 6.69TAKK209 pKa = 9.05VGPVEE214 pKa = 3.8GSVYY218 pKa = 10.49VRR220 pKa = 11.84VHH222 pKa = 5.11SAKK225 pKa = 10.18GRR227 pKa = 11.84NNTSVGRR234 pKa = 11.84LTLDD238 pKa = 2.98SVTFGLLVYY247 pKa = 10.53DD248 pKa = 4.7GSTVAGFSGGAYY260 pKa = 9.25YY261 pKa = 10.71LANRR265 pKa = 11.84VVGIHH270 pKa = 6.55CGGGAINYY278 pKa = 7.26GYY280 pKa = 10.78SMEE283 pKa = 4.48WIMSVLARR291 pKa = 11.84RR292 pKa = 11.84GSVPEE297 pKa = 4.42AGGQYY302 pKa = 11.18ALRR305 pKa = 11.84NILRR309 pKa = 11.84SARR312 pKa = 11.84KK313 pKa = 9.24SDD315 pKa = 3.54VQVTSSGNPDD325 pKa = 3.13EE326 pKa = 4.45VQVRR330 pKa = 11.84VGGKK334 pKa = 7.97FWIVDD339 pKa = 3.49RR340 pKa = 11.84EE341 pKa = 4.31DD342 pKa = 3.94YY343 pKa = 10.98VDD345 pKa = 5.34LMEE348 pKa = 5.77DD349 pKa = 4.67DD350 pKa = 3.58KK351 pKa = 11.43FGWMFDD357 pKa = 3.65DD358 pKa = 4.43DD359 pKa = 4.44SEE361 pKa = 5.12RR362 pKa = 11.84EE363 pKa = 3.83EE364 pKa = 4.37RR365 pKa = 11.84NMRR368 pKa = 11.84KK369 pKa = 9.57RR370 pKa = 11.84FRR372 pKa = 11.84DD373 pKa = 3.22TFYY376 pKa = 11.19EE377 pKa = 3.88NDD379 pKa = 3.52RR380 pKa = 11.84EE381 pKa = 4.41AQGEE385 pKa = 4.37LFPEE389 pKa = 5.15GSVVSTLNGSATPSGTQEE407 pKa = 3.97LQGLIQSQIQNTIEE421 pKa = 3.93IQSLKK426 pKa = 10.51SQQQVMNGLEE436 pKa = 4.28QKK438 pKa = 11.03LEE440 pKa = 4.09TLDD443 pKa = 4.08DD444 pKa = 4.32LLRR447 pKa = 11.84SIPITGHH454 pKa = 6.09APLMDD459 pKa = 3.45MLEE462 pKa = 4.06SLEE465 pKa = 4.26RR466 pKa = 11.84KK467 pKa = 7.85VTSLSQSLSRR477 pKa = 11.84QTSSPLRR484 pKa = 11.84EE485 pKa = 4.62DD486 pKa = 3.74LSQVRR491 pKa = 11.84TVGNSTSLPCSEE503 pKa = 4.92PSTSRR508 pKa = 11.84VQPVSVNSKK517 pKa = 8.47RR518 pKa = 11.84TTPSVRR524 pKa = 11.84PWDD527 pKa = 3.69GMEE530 pKa = 4.11SDD532 pKa = 3.84LRR534 pKa = 11.84TYY536 pKa = 7.23VTWRR540 pKa = 11.84SSVDD544 pKa = 2.98ASRR547 pKa = 11.84KK548 pKa = 9.72DD549 pKa = 3.96FYY551 pKa = 11.73ALRR554 pKa = 11.84TEE556 pKa = 4.51FLVQHH561 pKa = 6.15MPHH564 pKa = 5.96LTIEE568 pKa = 4.07QRR570 pKa = 11.84AEE572 pKa = 3.81LVRR575 pKa = 11.84LEE577 pKa = 3.9KK578 pKa = 10.54ARR580 pKa = 11.84RR581 pKa = 11.84VNRR584 pKa = 11.84KK585 pKa = 7.91KK586 pKa = 8.01TQKK589 pKa = 10.17KK590 pKa = 9.72SSEE593 pKa = 3.79GKK595 pKa = 8.78QRR597 pKa = 11.84KK598 pKa = 5.78EE599 pKa = 3.47QNRR602 pKa = 11.84AIQQTLSSLL611 pKa = 3.68
MM1 pKa = 7.08YY2 pKa = 10.2AVSMYY7 pKa = 10.86LATLLVWAVFMTYY20 pKa = 8.78TVVDD24 pKa = 5.21FFWDD28 pKa = 3.52WPVVRR33 pKa = 11.84QAVLRR38 pKa = 11.84DD39 pKa = 3.51RR40 pKa = 11.84AGKK43 pKa = 10.29ALFDD47 pKa = 3.86FCLGAVVLGIGFVLFFSSHH66 pKa = 7.82DD67 pKa = 3.24ILEE70 pKa = 4.62WIGRR74 pKa = 11.84GVEE77 pKa = 4.06TLVRR81 pKa = 11.84RR82 pKa = 11.84IWKK85 pKa = 9.4FCFARR90 pKa = 11.84ATARR94 pKa = 11.84VARR97 pKa = 11.84PDD99 pKa = 2.92KK100 pKa = 10.95SMRR103 pKa = 11.84RR104 pKa = 11.84VLEE107 pKa = 4.06TMKK110 pKa = 10.6EE111 pKa = 4.17GSDD114 pKa = 3.51LQTIPRR120 pKa = 11.84PACQLALYY128 pKa = 9.8DD129 pKa = 3.74YY130 pKa = 11.35DD131 pKa = 4.32SIEE134 pKa = 3.91RR135 pKa = 11.84QKK137 pKa = 11.25YY138 pKa = 7.83YY139 pKa = 10.73LGGAIHH145 pKa = 6.77VEE147 pKa = 4.24GGWLVSCTHH156 pKa = 6.7IVNQLRR162 pKa = 11.84NKK164 pKa = 9.76PYY166 pKa = 10.94ASVLDD171 pKa = 4.09PQGVEE176 pKa = 3.52QFYY179 pKa = 11.02SLEE182 pKa = 3.95GLEE185 pKa = 4.15WEE187 pKa = 4.86EE188 pKa = 3.84IGADD192 pKa = 3.45VSVAKK197 pKa = 10.55LGRR200 pKa = 11.84VFQHH204 pKa = 6.62LKK206 pKa = 6.69TAKK209 pKa = 9.05VGPVEE214 pKa = 3.8GSVYY218 pKa = 10.49VRR220 pKa = 11.84VHH222 pKa = 5.11SAKK225 pKa = 10.18GRR227 pKa = 11.84NNTSVGRR234 pKa = 11.84LTLDD238 pKa = 2.98SVTFGLLVYY247 pKa = 10.53DD248 pKa = 4.7GSTVAGFSGGAYY260 pKa = 9.25YY261 pKa = 10.71LANRR265 pKa = 11.84VVGIHH270 pKa = 6.55CGGGAINYY278 pKa = 7.26GYY280 pKa = 10.78SMEE283 pKa = 4.48WIMSVLARR291 pKa = 11.84RR292 pKa = 11.84GSVPEE297 pKa = 4.42AGGQYY302 pKa = 11.18ALRR305 pKa = 11.84NILRR309 pKa = 11.84SARR312 pKa = 11.84KK313 pKa = 9.24SDD315 pKa = 3.54VQVTSSGNPDD325 pKa = 3.13EE326 pKa = 4.45VQVRR330 pKa = 11.84VGGKK334 pKa = 7.97FWIVDD339 pKa = 3.49RR340 pKa = 11.84EE341 pKa = 4.31DD342 pKa = 3.94YY343 pKa = 10.98VDD345 pKa = 5.34LMEE348 pKa = 5.77DD349 pKa = 4.67DD350 pKa = 3.58KK351 pKa = 11.43FGWMFDD357 pKa = 3.65DD358 pKa = 4.43DD359 pKa = 4.44SEE361 pKa = 5.12RR362 pKa = 11.84EE363 pKa = 3.83EE364 pKa = 4.37RR365 pKa = 11.84NMRR368 pKa = 11.84KK369 pKa = 9.57RR370 pKa = 11.84FRR372 pKa = 11.84DD373 pKa = 3.22TFYY376 pKa = 11.19EE377 pKa = 3.88NDD379 pKa = 3.52RR380 pKa = 11.84EE381 pKa = 4.41AQGEE385 pKa = 4.37LFPEE389 pKa = 5.15GSVVSTLNGSATPSGTQEE407 pKa = 3.97LQGLIQSQIQNTIEE421 pKa = 3.93IQSLKK426 pKa = 10.51SQQQVMNGLEE436 pKa = 4.28QKK438 pKa = 11.03LEE440 pKa = 4.09TLDD443 pKa = 4.08DD444 pKa = 4.32LLRR447 pKa = 11.84SIPITGHH454 pKa = 6.09APLMDD459 pKa = 3.45MLEE462 pKa = 4.06SLEE465 pKa = 4.26RR466 pKa = 11.84KK467 pKa = 7.85VTSLSQSLSRR477 pKa = 11.84QTSSPLRR484 pKa = 11.84EE485 pKa = 4.62DD486 pKa = 3.74LSQVRR491 pKa = 11.84TVGNSTSLPCSEE503 pKa = 4.92PSTSRR508 pKa = 11.84VQPVSVNSKK517 pKa = 8.47RR518 pKa = 11.84TTPSVRR524 pKa = 11.84PWDD527 pKa = 3.69GMEE530 pKa = 4.11SDD532 pKa = 3.84LRR534 pKa = 11.84TYY536 pKa = 7.23VTWRR540 pKa = 11.84SSVDD544 pKa = 2.98ASRR547 pKa = 11.84KK548 pKa = 9.72DD549 pKa = 3.96FYY551 pKa = 11.73ALRR554 pKa = 11.84TEE556 pKa = 4.51FLVQHH561 pKa = 6.15MPHH564 pKa = 5.96LTIEE568 pKa = 4.07QRR570 pKa = 11.84AEE572 pKa = 3.81LVRR575 pKa = 11.84LEE577 pKa = 3.9KK578 pKa = 10.54ARR580 pKa = 11.84RR581 pKa = 11.84VNRR584 pKa = 11.84KK585 pKa = 7.91KK586 pKa = 8.01TQKK589 pKa = 10.17KK590 pKa = 9.72SSEE593 pKa = 3.79GKK595 pKa = 8.78QRR597 pKa = 11.84KK598 pKa = 5.78EE599 pKa = 3.47QNRR602 pKa = 11.84AIQQTLSSLL611 pKa = 3.68
Molecular weight: 68.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
832 |
221 |
611 |
416.0 |
47.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 0.738 | 1.563 ± 0.821 |
5.048 ± 0.499 | 6.37 ± 0.018 |
3.726 ± 0.177 | 7.332 ± 0.279 |
1.683 ± 0.297 | 2.885 ± 1.013 |
4.087 ± 0.239 | 10.096 ± 1.086 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.486 ± 1.458 | 2.524 ± 0.365 |
3.726 ± 0.64 | 4.808 ± 0.84 |
8.053 ± 0.416 | 8.293 ± 1.234 |
4.808 ± 1.071 | 9.014 ± 0.677 |
2.524 ± 0.792 | 3.726 ± 0.64 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
