
Sclerotinia sclerotiorum partitivirus S
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
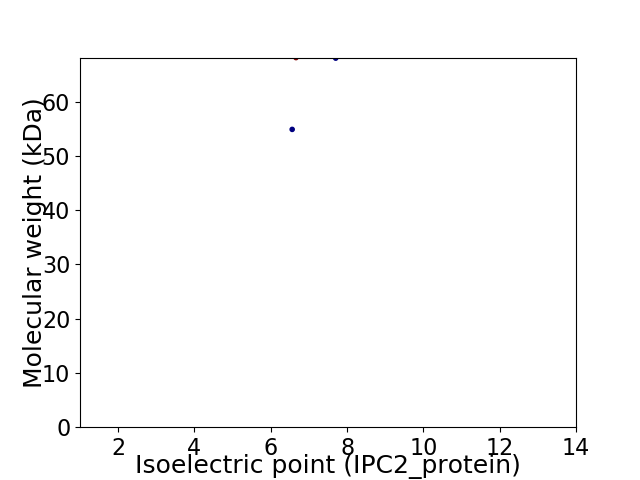
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
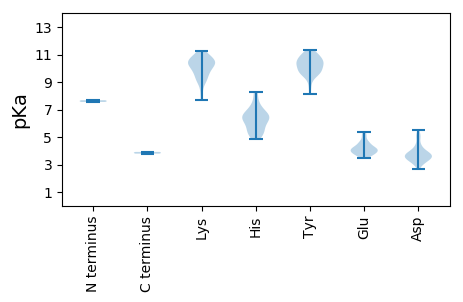
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|C7ENF5|C7ENF5_9VIRU Putative coat protein OS=Sclerotinia sclerotiorum partitivirus S OX=659497 PE=4 SV=1
MM1 pKa = 7.55SSRR4 pKa = 11.84SARR7 pKa = 11.84KK8 pKa = 9.7VSTQEE13 pKa = 3.83KK14 pKa = 9.23TSSQSGKK21 pKa = 9.71NRR23 pKa = 11.84ASKK26 pKa = 10.79KK27 pKa = 9.38SGKK30 pKa = 9.85KK31 pKa = 9.31SAPPEE36 pKa = 4.06YY37 pKa = 10.7SVDD40 pKa = 3.84TPSEE44 pKa = 4.01QSDD47 pKa = 4.05VEE49 pKa = 4.17SDD51 pKa = 3.01ILSEE55 pKa = 5.06LEE57 pKa = 4.37VSDD60 pKa = 5.54ADD62 pKa = 3.94DD63 pKa = 3.74ASRR66 pKa = 11.84PVPASKK72 pKa = 10.35SKK74 pKa = 10.82KK75 pKa = 8.12GTTRR79 pKa = 11.84NKK81 pKa = 10.3KK82 pKa = 9.6KK83 pKa = 9.57EE84 pKa = 3.85KK85 pKa = 10.4SVASSAGSSIPPYY98 pKa = 10.95MMMLTALNFFSLTEE112 pKa = 4.06IEE114 pKa = 4.72HH115 pKa = 5.12VTISTYY121 pKa = 10.4TPSCWSMYY129 pKa = 10.62AVLDD133 pKa = 3.97AMHH136 pKa = 7.32DD137 pKa = 3.87LVGDD141 pKa = 3.7NASLRR146 pKa = 11.84RR147 pKa = 11.84FCPYY151 pKa = 10.49YY152 pKa = 10.58HH153 pKa = 6.48VALSNIYY160 pKa = 9.99YY161 pKa = 10.3GIIFIIQVLRR171 pKa = 11.84ANQVANNLSQADD183 pKa = 3.85FQFLRR188 pKa = 11.84FFEE191 pKa = 5.27SNFALEE197 pKa = 4.59EE198 pKa = 4.21LPVAGPLVLFFQNLAAFKK216 pKa = 10.35PDD218 pKa = 3.0GNRR221 pKa = 11.84FNWVVPHH228 pKa = 5.56YY229 pKa = 10.99NNYY232 pKa = 9.86GPANGANSPRR242 pKa = 11.84VNAQSTALPQLPQMISLLNLFGASNAALLTAMDD275 pKa = 4.39TAGQWEE281 pKa = 5.12PFTFAAGGTIAGFAYY296 pKa = 10.26GAGFATDD303 pKa = 4.31AAASDD308 pKa = 3.58QTLRR312 pKa = 11.84APGINVPWQHH322 pKa = 6.47DD323 pKa = 3.73TAIHH327 pKa = 6.29RR328 pKa = 11.84KK329 pKa = 8.82LVPISRR335 pKa = 11.84RR336 pKa = 11.84MSVPTMNGAKK346 pKa = 10.11DD347 pKa = 3.58SPRR350 pKa = 11.84TYY352 pKa = 10.52CGLDD356 pKa = 3.47GQFHH360 pKa = 6.91WFHH363 pKa = 6.53KK364 pKa = 10.81AIGAVSQEE372 pKa = 3.77AKK374 pKa = 10.45YY375 pKa = 9.94FAGSTTLANINPSTGPSAVVEE396 pKa = 4.42TTVAVPAAAIPQATAWYY413 pKa = 9.08EE414 pKa = 4.07GYY416 pKa = 9.8PFNLNLTVNTRR427 pKa = 11.84VRR429 pKa = 11.84SISEE433 pKa = 3.87TDD435 pKa = 3.3VKK437 pKa = 10.74IGTFAATCNIITGGNFTRR455 pKa = 11.84WNGSSVFAGDD465 pKa = 3.57FFVAATNPINFEE477 pKa = 3.91NPTEE481 pKa = 4.16VNAIAMAQPTMTLSLYY497 pKa = 10.05RR498 pKa = 11.84PKK500 pKa = 10.82GGEE503 pKa = 4.06ANSDD507 pKa = 3.7DD508 pKa = 3.79
MM1 pKa = 7.55SSRR4 pKa = 11.84SARR7 pKa = 11.84KK8 pKa = 9.7VSTQEE13 pKa = 3.83KK14 pKa = 9.23TSSQSGKK21 pKa = 9.71NRR23 pKa = 11.84ASKK26 pKa = 10.79KK27 pKa = 9.38SGKK30 pKa = 9.85KK31 pKa = 9.31SAPPEE36 pKa = 4.06YY37 pKa = 10.7SVDD40 pKa = 3.84TPSEE44 pKa = 4.01QSDD47 pKa = 4.05VEE49 pKa = 4.17SDD51 pKa = 3.01ILSEE55 pKa = 5.06LEE57 pKa = 4.37VSDD60 pKa = 5.54ADD62 pKa = 3.94DD63 pKa = 3.74ASRR66 pKa = 11.84PVPASKK72 pKa = 10.35SKK74 pKa = 10.82KK75 pKa = 8.12GTTRR79 pKa = 11.84NKK81 pKa = 10.3KK82 pKa = 9.6KK83 pKa = 9.57EE84 pKa = 3.85KK85 pKa = 10.4SVASSAGSSIPPYY98 pKa = 10.95MMMLTALNFFSLTEE112 pKa = 4.06IEE114 pKa = 4.72HH115 pKa = 5.12VTISTYY121 pKa = 10.4TPSCWSMYY129 pKa = 10.62AVLDD133 pKa = 3.97AMHH136 pKa = 7.32DD137 pKa = 3.87LVGDD141 pKa = 3.7NASLRR146 pKa = 11.84RR147 pKa = 11.84FCPYY151 pKa = 10.49YY152 pKa = 10.58HH153 pKa = 6.48VALSNIYY160 pKa = 9.99YY161 pKa = 10.3GIIFIIQVLRR171 pKa = 11.84ANQVANNLSQADD183 pKa = 3.85FQFLRR188 pKa = 11.84FFEE191 pKa = 5.27SNFALEE197 pKa = 4.59EE198 pKa = 4.21LPVAGPLVLFFQNLAAFKK216 pKa = 10.35PDD218 pKa = 3.0GNRR221 pKa = 11.84FNWVVPHH228 pKa = 5.56YY229 pKa = 10.99NNYY232 pKa = 9.86GPANGANSPRR242 pKa = 11.84VNAQSTALPQLPQMISLLNLFGASNAALLTAMDD275 pKa = 4.39TAGQWEE281 pKa = 5.12PFTFAAGGTIAGFAYY296 pKa = 10.26GAGFATDD303 pKa = 4.31AAASDD308 pKa = 3.58QTLRR312 pKa = 11.84APGINVPWQHH322 pKa = 6.47DD323 pKa = 3.73TAIHH327 pKa = 6.29RR328 pKa = 11.84KK329 pKa = 8.82LVPISRR335 pKa = 11.84RR336 pKa = 11.84MSVPTMNGAKK346 pKa = 10.11DD347 pKa = 3.58SPRR350 pKa = 11.84TYY352 pKa = 10.52CGLDD356 pKa = 3.47GQFHH360 pKa = 6.91WFHH363 pKa = 6.53KK364 pKa = 10.81AIGAVSQEE372 pKa = 3.77AKK374 pKa = 10.45YY375 pKa = 9.94FAGSTTLANINPSTGPSAVVEE396 pKa = 4.42TTVAVPAAAIPQATAWYY413 pKa = 9.08EE414 pKa = 4.07GYY416 pKa = 9.8PFNLNLTVNTRR427 pKa = 11.84VRR429 pKa = 11.84SISEE433 pKa = 3.87TDD435 pKa = 3.3VKK437 pKa = 10.74IGTFAATCNIITGGNFTRR455 pKa = 11.84WNGSSVFAGDD465 pKa = 3.57FFVAATNPINFEE477 pKa = 3.91NPTEE481 pKa = 4.16VNAIAMAQPTMTLSLYY497 pKa = 10.05RR498 pKa = 11.84PKK500 pKa = 10.82GGEE503 pKa = 4.06ANSDD507 pKa = 3.7DD508 pKa = 3.79
Molecular weight: 54.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7ENF5|C7ENF5_9VIRU Putative coat protein OS=Sclerotinia sclerotiorum partitivirus S OX=659497 PE=4 SV=1
MM1 pKa = 7.68PALNLRR7 pKa = 11.84YY8 pKa = 9.4LFSISRR14 pKa = 11.84SEE16 pKa = 3.85MKK18 pKa = 10.3KK19 pKa = 9.69IRR21 pKa = 11.84SSHH24 pKa = 5.18TKK26 pKa = 9.07VSPRR30 pKa = 11.84EE31 pKa = 3.61EE32 pKa = 3.76ATRR35 pKa = 11.84FSILKK40 pKa = 9.8HH41 pKa = 6.48AILKK45 pKa = 9.76HH46 pKa = 5.58GSVGLLNQVLLGKK59 pKa = 10.02RR60 pKa = 11.84RR61 pKa = 11.84SDD63 pKa = 3.36ASDD66 pKa = 2.95EE67 pKa = 4.29RR68 pKa = 11.84LIQDD72 pKa = 3.18FHH74 pKa = 8.13EE75 pKa = 5.12FEE77 pKa = 4.59QPVHH81 pKa = 5.79PVPRR85 pKa = 11.84DD86 pKa = 3.36KK87 pKa = 10.99HH88 pKa = 4.88YY89 pKa = 11.29LRR91 pKa = 11.84ALRR94 pKa = 11.84VTEE97 pKa = 4.57KK98 pKa = 10.77LMKK101 pKa = 9.23PAKK104 pKa = 8.33TLHH107 pKa = 6.74PISFPDD113 pKa = 3.35LRR115 pKa = 11.84YY116 pKa = 10.26YY117 pKa = 10.29PWTKK121 pKa = 10.41NVSAEE126 pKa = 3.73APYY129 pKa = 10.92NFEE132 pKa = 4.16KK133 pKa = 10.53KK134 pKa = 10.23YY135 pKa = 11.18EE136 pKa = 3.92EE137 pKa = 4.28LLRR140 pKa = 11.84DD141 pKa = 3.66KK142 pKa = 11.01QRR144 pKa = 11.84LGEE147 pKa = 4.29IEE149 pKa = 3.98TATATFHH156 pKa = 6.47NLEE159 pKa = 4.55DD160 pKa = 5.21EE161 pKa = 4.14IFEE164 pKa = 4.69DD165 pKa = 4.39NRR167 pKa = 11.84YY168 pKa = 9.74LIHH171 pKa = 7.18KK172 pKa = 9.06IKK174 pKa = 10.88EE175 pKa = 4.01GDD177 pKa = 3.61SQFWDD182 pKa = 3.25KK183 pKa = 11.26DD184 pKa = 3.75GKK186 pKa = 9.32PRR188 pKa = 11.84PYY190 pKa = 11.05YY191 pKa = 9.64HH192 pKa = 6.36TTLHH196 pKa = 6.06ARR198 pKa = 11.84AHH200 pKa = 5.21VVGHH204 pKa = 6.56EE205 pKa = 4.23DD206 pKa = 3.15ADD208 pKa = 4.06KK209 pKa = 10.37IRR211 pKa = 11.84AVFGVPKK218 pKa = 10.18LLLMAEE224 pKa = 4.52NMFIWPLQAYY234 pKa = 8.11YY235 pKa = 11.04LNQDD239 pKa = 3.54TTKK242 pKa = 10.59HH243 pKa = 5.45HH244 pKa = 6.57LLWGNEE250 pKa = 3.5IMKK253 pKa = 10.57GGWKK257 pKa = 10.03KK258 pKa = 10.68LWGQLQNGRR267 pKa = 11.84ISRR270 pKa = 11.84TILSLDD276 pKa = 2.65WSEE279 pKa = 3.76FDD281 pKa = 4.32KK282 pKa = 11.22RR283 pKa = 11.84ALHH286 pKa = 6.1EE287 pKa = 4.76VIDD290 pKa = 5.01DD291 pKa = 3.69VHH293 pKa = 8.26SMWKK297 pKa = 9.05SWFDD301 pKa = 3.42FTHH304 pKa = 6.08YY305 pKa = 10.45EE306 pKa = 3.6PTIFYY311 pKa = 9.59EE312 pKa = 4.28KK313 pKa = 11.19GEE315 pKa = 4.02IPEE318 pKa = 3.68PHH320 pKa = 6.54RR321 pKa = 11.84RR322 pKa = 11.84IEE324 pKa = 4.17NLWIWMTDD332 pKa = 3.28MVKK335 pKa = 9.96HH336 pKa = 5.24YY337 pKa = 9.92PILQPDD343 pKa = 3.95GKK345 pKa = 10.64VYY347 pKa = 9.72QWTRR351 pKa = 11.84NGIASGFQQTQLLDD365 pKa = 3.61SFVNMIMLLTVLSANGINIEE385 pKa = 5.4HH386 pKa = 7.44PDD388 pKa = 3.43FWIKK392 pKa = 10.53VQGDD396 pKa = 3.66DD397 pKa = 3.6SLISIVEE404 pKa = 3.83RR405 pKa = 11.84RR406 pKa = 11.84FQMFGISYY414 pKa = 10.69LDD416 pKa = 3.46TLADD420 pKa = 3.6LASYY424 pKa = 9.58YY425 pKa = 10.79FNAKK429 pKa = 10.29LSVKK433 pKa = 10.17KK434 pKa = 10.76SFISDD439 pKa = 3.81TPQGQYY445 pKa = 10.31VLGYY449 pKa = 8.67FNHH452 pKa = 6.15YY453 pKa = 9.49GIPYY457 pKa = 10.38RR458 pKa = 11.84LDD460 pKa = 4.73DD461 pKa = 5.11DD462 pKa = 5.05LLSHH466 pKa = 6.86LVFPEE471 pKa = 4.1RR472 pKa = 11.84PQRR475 pKa = 11.84LEE477 pKa = 3.63EE478 pKa = 4.02TAASCVGIAMASMGCSKK495 pKa = 10.42VVYY498 pKa = 9.41SICDD502 pKa = 3.33DD503 pKa = 3.88AYY505 pKa = 9.72TFITKK510 pKa = 7.67TLRR513 pKa = 11.84RR514 pKa = 11.84PAKK517 pKa = 10.18AGSLFWLEE525 pKa = 3.71RR526 pKa = 11.84AFGYY530 pKa = 9.98EE531 pKa = 3.85MPNIAKK537 pKa = 8.89MPTFEE542 pKa = 4.01EE543 pKa = 4.29CLYY546 pKa = 10.9ASYY549 pKa = 10.19DD550 pKa = 3.36IPVRR554 pKa = 11.84TEE556 pKa = 3.48NMKK559 pKa = 10.32QRR561 pKa = 11.84LWPTNAKK568 pKa = 9.76AKK570 pKa = 10.34NGFYY574 pKa = 10.55FLRR577 pKa = 11.84HH578 pKa = 5.46LCC580 pKa = 3.92
MM1 pKa = 7.68PALNLRR7 pKa = 11.84YY8 pKa = 9.4LFSISRR14 pKa = 11.84SEE16 pKa = 3.85MKK18 pKa = 10.3KK19 pKa = 9.69IRR21 pKa = 11.84SSHH24 pKa = 5.18TKK26 pKa = 9.07VSPRR30 pKa = 11.84EE31 pKa = 3.61EE32 pKa = 3.76ATRR35 pKa = 11.84FSILKK40 pKa = 9.8HH41 pKa = 6.48AILKK45 pKa = 9.76HH46 pKa = 5.58GSVGLLNQVLLGKK59 pKa = 10.02RR60 pKa = 11.84RR61 pKa = 11.84SDD63 pKa = 3.36ASDD66 pKa = 2.95EE67 pKa = 4.29RR68 pKa = 11.84LIQDD72 pKa = 3.18FHH74 pKa = 8.13EE75 pKa = 5.12FEE77 pKa = 4.59QPVHH81 pKa = 5.79PVPRR85 pKa = 11.84DD86 pKa = 3.36KK87 pKa = 10.99HH88 pKa = 4.88YY89 pKa = 11.29LRR91 pKa = 11.84ALRR94 pKa = 11.84VTEE97 pKa = 4.57KK98 pKa = 10.77LMKK101 pKa = 9.23PAKK104 pKa = 8.33TLHH107 pKa = 6.74PISFPDD113 pKa = 3.35LRR115 pKa = 11.84YY116 pKa = 10.26YY117 pKa = 10.29PWTKK121 pKa = 10.41NVSAEE126 pKa = 3.73APYY129 pKa = 10.92NFEE132 pKa = 4.16KK133 pKa = 10.53KK134 pKa = 10.23YY135 pKa = 11.18EE136 pKa = 3.92EE137 pKa = 4.28LLRR140 pKa = 11.84DD141 pKa = 3.66KK142 pKa = 11.01QRR144 pKa = 11.84LGEE147 pKa = 4.29IEE149 pKa = 3.98TATATFHH156 pKa = 6.47NLEE159 pKa = 4.55DD160 pKa = 5.21EE161 pKa = 4.14IFEE164 pKa = 4.69DD165 pKa = 4.39NRR167 pKa = 11.84YY168 pKa = 9.74LIHH171 pKa = 7.18KK172 pKa = 9.06IKK174 pKa = 10.88EE175 pKa = 4.01GDD177 pKa = 3.61SQFWDD182 pKa = 3.25KK183 pKa = 11.26DD184 pKa = 3.75GKK186 pKa = 9.32PRR188 pKa = 11.84PYY190 pKa = 11.05YY191 pKa = 9.64HH192 pKa = 6.36TTLHH196 pKa = 6.06ARR198 pKa = 11.84AHH200 pKa = 5.21VVGHH204 pKa = 6.56EE205 pKa = 4.23DD206 pKa = 3.15ADD208 pKa = 4.06KK209 pKa = 10.37IRR211 pKa = 11.84AVFGVPKK218 pKa = 10.18LLLMAEE224 pKa = 4.52NMFIWPLQAYY234 pKa = 8.11YY235 pKa = 11.04LNQDD239 pKa = 3.54TTKK242 pKa = 10.59HH243 pKa = 5.45HH244 pKa = 6.57LLWGNEE250 pKa = 3.5IMKK253 pKa = 10.57GGWKK257 pKa = 10.03KK258 pKa = 10.68LWGQLQNGRR267 pKa = 11.84ISRR270 pKa = 11.84TILSLDD276 pKa = 2.65WSEE279 pKa = 3.76FDD281 pKa = 4.32KK282 pKa = 11.22RR283 pKa = 11.84ALHH286 pKa = 6.1EE287 pKa = 4.76VIDD290 pKa = 5.01DD291 pKa = 3.69VHH293 pKa = 8.26SMWKK297 pKa = 9.05SWFDD301 pKa = 3.42FTHH304 pKa = 6.08YY305 pKa = 10.45EE306 pKa = 3.6PTIFYY311 pKa = 9.59EE312 pKa = 4.28KK313 pKa = 11.19GEE315 pKa = 4.02IPEE318 pKa = 3.68PHH320 pKa = 6.54RR321 pKa = 11.84RR322 pKa = 11.84IEE324 pKa = 4.17NLWIWMTDD332 pKa = 3.28MVKK335 pKa = 9.96HH336 pKa = 5.24YY337 pKa = 9.92PILQPDD343 pKa = 3.95GKK345 pKa = 10.64VYY347 pKa = 9.72QWTRR351 pKa = 11.84NGIASGFQQTQLLDD365 pKa = 3.61SFVNMIMLLTVLSANGINIEE385 pKa = 5.4HH386 pKa = 7.44PDD388 pKa = 3.43FWIKK392 pKa = 10.53VQGDD396 pKa = 3.66DD397 pKa = 3.6SLISIVEE404 pKa = 3.83RR405 pKa = 11.84RR406 pKa = 11.84FQMFGISYY414 pKa = 10.69LDD416 pKa = 3.46TLADD420 pKa = 3.6LASYY424 pKa = 9.58YY425 pKa = 10.79FNAKK429 pKa = 10.29LSVKK433 pKa = 10.17KK434 pKa = 10.76SFISDD439 pKa = 3.81TPQGQYY445 pKa = 10.31VLGYY449 pKa = 8.67FNHH452 pKa = 6.15YY453 pKa = 9.49GIPYY457 pKa = 10.38RR458 pKa = 11.84LDD460 pKa = 4.73DD461 pKa = 5.11DD462 pKa = 5.05LLSHH466 pKa = 6.86LVFPEE471 pKa = 4.1RR472 pKa = 11.84PQRR475 pKa = 11.84LEE477 pKa = 3.63EE478 pKa = 4.02TAASCVGIAMASMGCSKK495 pKa = 10.42VVYY498 pKa = 9.41SICDD502 pKa = 3.33DD503 pKa = 3.88AYY505 pKa = 9.72TFITKK510 pKa = 7.67TLRR513 pKa = 11.84RR514 pKa = 11.84PAKK517 pKa = 10.18AGSLFWLEE525 pKa = 3.71RR526 pKa = 11.84AFGYY530 pKa = 9.98EE531 pKa = 3.85MPNIAKK537 pKa = 8.89MPTFEE542 pKa = 4.01EE543 pKa = 4.29CLYY546 pKa = 10.9ASYY549 pKa = 10.19DD550 pKa = 3.36IPVRR554 pKa = 11.84TEE556 pKa = 3.48NMKK559 pKa = 10.32QRR561 pKa = 11.84LWPTNAKK568 pKa = 9.76AKK570 pKa = 10.34NGFYY574 pKa = 10.55FLRR577 pKa = 11.84HH578 pKa = 5.46LCC580 pKa = 3.92
Molecular weight: 68.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1088 |
508 |
580 |
544.0 |
61.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.824 ± 2.218 | 0.827 ± 0.026 |
5.055 ± 0.604 | 5.239 ± 0.854 |
5.331 ± 0.119 | 5.515 ± 0.515 |
2.941 ± 0.896 | 5.423 ± 0.587 |
5.607 ± 0.966 | 8.272 ± 1.294 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.665 ± 0.199 | 5.055 ± 1.074 |
5.607 ± 0.454 | 3.493 ± 0.033 |
5.055 ± 0.733 | 7.813 ± 1.332 |
5.974 ± 0.859 | 5.239 ± 0.566 |
2.022 ± 0.422 | 4.044 ± 0.587 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
