
Huangpi Tick Virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
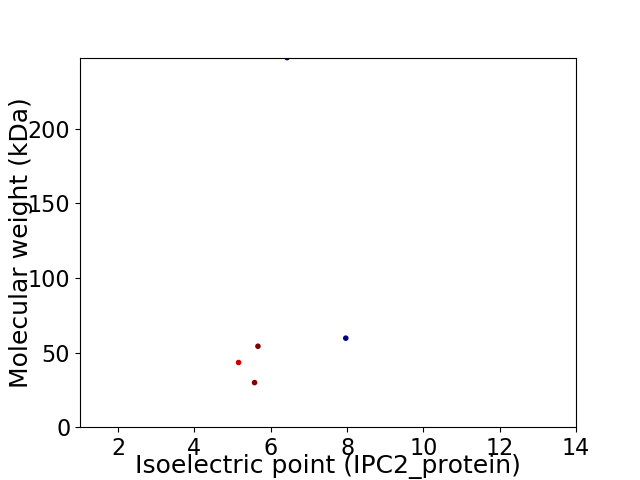
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KK14|A0A0B5KK14_9RHAB Putative matrix protein OS=Huangpi Tick Virus 3 OX=1608049 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.22QRR3 pKa = 11.84QLPTLPDD10 pKa = 3.52HH11 pKa = 6.61VVEE14 pKa = 4.35ALRR17 pKa = 11.84DD18 pKa = 3.55LSEE21 pKa = 4.02EE22 pKa = 3.9RR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84DD27 pKa = 3.42PLGLEE32 pKa = 3.92DD33 pKa = 3.66HH34 pKa = 7.05LEE36 pKa = 3.96RR37 pKa = 11.84SAYY40 pKa = 10.24RR41 pKa = 11.84RR42 pKa = 11.84MSASPARR49 pKa = 11.84MEE51 pKa = 4.32TEE53 pKa = 4.23ADD55 pKa = 3.28QDD57 pKa = 4.05PAPSGTPKK65 pKa = 10.49KK66 pKa = 9.87PRR68 pKa = 11.84KK69 pKa = 8.24QTGQDD74 pKa = 3.13KK75 pKa = 9.62PHH77 pKa = 7.14AGEE80 pKa = 4.02GARR83 pKa = 11.84LLRR86 pKa = 11.84STSKK90 pKa = 10.22QRR92 pKa = 11.84PEE94 pKa = 4.18VIGKK98 pKa = 9.75RR99 pKa = 11.84SKK101 pKa = 10.42ATDD104 pKa = 3.15RR105 pKa = 11.84PQRR108 pKa = 11.84SNVSMKK114 pKa = 10.47QGANQKK120 pKa = 9.72GGEE123 pKa = 4.24VSVQTHH129 pKa = 5.42TTPDD133 pKa = 3.63PSDD136 pKa = 3.55EE137 pKa = 4.33PTSEE141 pKa = 3.89MKK143 pKa = 10.52EE144 pKa = 4.05PNIGAEE150 pKa = 4.14GVVVLTEE157 pKa = 4.1PVPDD161 pKa = 5.09DD162 pKa = 4.82VIDD165 pKa = 4.2DD166 pKa = 3.53QMEE169 pKa = 4.07IDD171 pKa = 4.11KK172 pKa = 10.66QDD174 pKa = 3.64PSTNPQEE181 pKa = 4.48TYY183 pKa = 10.55FVATTSIFDD192 pKa = 3.95PADD195 pKa = 3.44LSGEE199 pKa = 4.31SLTAPQTLALQKK211 pKa = 10.58CALCMEE217 pKa = 4.69NLLAHH222 pKa = 6.95GFAKK226 pKa = 10.59DD227 pKa = 3.13ISADD231 pKa = 3.38VSNQGIVLRR240 pKa = 11.84YY241 pKa = 7.9TRR243 pKa = 11.84LLPAFDD249 pKa = 4.42TPPGEE254 pKa = 4.43QTCTQPPATDD264 pKa = 3.6PASAHH269 pKa = 5.92QKK271 pKa = 9.78SQDD274 pKa = 3.45KK275 pKa = 10.56QGPPDD280 pKa = 4.04KK281 pKa = 9.78EE282 pKa = 4.14APPAPTSDD290 pKa = 3.91KK291 pKa = 11.04NLEE294 pKa = 4.25PPLDD298 pKa = 4.43DD299 pKa = 5.75DD300 pKa = 3.98EE301 pKa = 4.88QFARR305 pKa = 11.84QVYY308 pKa = 9.54RR309 pKa = 11.84ALQDD313 pKa = 3.15MTIEE317 pKa = 4.3VPSRR321 pKa = 11.84SPGQMLSVKK330 pKa = 10.03VSAVPLRR337 pKa = 11.84WSVVWDD343 pKa = 3.26YY344 pKa = 11.83LKK346 pKa = 11.15AHH348 pKa = 6.23NRR350 pKa = 11.84VNWFIDD356 pKa = 3.72SASDD360 pKa = 3.6PAGKK364 pKa = 9.68LAFCAWSCRR373 pKa = 11.84VPSLMNTISPVFIRR387 pKa = 11.84DD388 pKa = 3.84AIKK391 pKa = 10.42GAPVV395 pKa = 2.82
MM1 pKa = 7.22QRR3 pKa = 11.84QLPTLPDD10 pKa = 3.52HH11 pKa = 6.61VVEE14 pKa = 4.35ALRR17 pKa = 11.84DD18 pKa = 3.55LSEE21 pKa = 4.02EE22 pKa = 3.9RR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84DD27 pKa = 3.42PLGLEE32 pKa = 3.92DD33 pKa = 3.66HH34 pKa = 7.05LEE36 pKa = 3.96RR37 pKa = 11.84SAYY40 pKa = 10.24RR41 pKa = 11.84RR42 pKa = 11.84MSASPARR49 pKa = 11.84MEE51 pKa = 4.32TEE53 pKa = 4.23ADD55 pKa = 3.28QDD57 pKa = 4.05PAPSGTPKK65 pKa = 10.49KK66 pKa = 9.87PRR68 pKa = 11.84KK69 pKa = 8.24QTGQDD74 pKa = 3.13KK75 pKa = 9.62PHH77 pKa = 7.14AGEE80 pKa = 4.02GARR83 pKa = 11.84LLRR86 pKa = 11.84STSKK90 pKa = 10.22QRR92 pKa = 11.84PEE94 pKa = 4.18VIGKK98 pKa = 9.75RR99 pKa = 11.84SKK101 pKa = 10.42ATDD104 pKa = 3.15RR105 pKa = 11.84PQRR108 pKa = 11.84SNVSMKK114 pKa = 10.47QGANQKK120 pKa = 9.72GGEE123 pKa = 4.24VSVQTHH129 pKa = 5.42TTPDD133 pKa = 3.63PSDD136 pKa = 3.55EE137 pKa = 4.33PTSEE141 pKa = 3.89MKK143 pKa = 10.52EE144 pKa = 4.05PNIGAEE150 pKa = 4.14GVVVLTEE157 pKa = 4.1PVPDD161 pKa = 5.09DD162 pKa = 4.82VIDD165 pKa = 4.2DD166 pKa = 3.53QMEE169 pKa = 4.07IDD171 pKa = 4.11KK172 pKa = 10.66QDD174 pKa = 3.64PSTNPQEE181 pKa = 4.48TYY183 pKa = 10.55FVATTSIFDD192 pKa = 3.95PADD195 pKa = 3.44LSGEE199 pKa = 4.31SLTAPQTLALQKK211 pKa = 10.58CALCMEE217 pKa = 4.69NLLAHH222 pKa = 6.95GFAKK226 pKa = 10.59DD227 pKa = 3.13ISADD231 pKa = 3.38VSNQGIVLRR240 pKa = 11.84YY241 pKa = 7.9TRR243 pKa = 11.84LLPAFDD249 pKa = 4.42TPPGEE254 pKa = 4.43QTCTQPPATDD264 pKa = 3.6PASAHH269 pKa = 5.92QKK271 pKa = 9.78SQDD274 pKa = 3.45KK275 pKa = 10.56QGPPDD280 pKa = 4.04KK281 pKa = 9.78EE282 pKa = 4.14APPAPTSDD290 pKa = 3.91KK291 pKa = 11.04NLEE294 pKa = 4.25PPLDD298 pKa = 4.43DD299 pKa = 5.75DD300 pKa = 3.98EE301 pKa = 4.88QFARR305 pKa = 11.84QVYY308 pKa = 9.54RR309 pKa = 11.84ALQDD313 pKa = 3.15MTIEE317 pKa = 4.3VPSRR321 pKa = 11.84SPGQMLSVKK330 pKa = 10.03VSAVPLRR337 pKa = 11.84WSVVWDD343 pKa = 3.26YY344 pKa = 11.83LKK346 pKa = 11.15AHH348 pKa = 6.23NRR350 pKa = 11.84VNWFIDD356 pKa = 3.72SASDD360 pKa = 3.6PAGKK364 pKa = 9.68LAFCAWSCRR373 pKa = 11.84VPSLMNTISPVFIRR387 pKa = 11.84DD388 pKa = 3.84AIKK391 pKa = 10.42GAPVV395 pKa = 2.82
Molecular weight: 43.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KTD9|A0A0B5KTD9_9RHAB GDP polyribonucleotidyltransferase OS=Huangpi Tick Virus 3 OX=1608049 GN=L PE=4 SV=1
MM1 pKa = 7.31RR2 pKa = 11.84TEE4 pKa = 4.47LCLSCLLLLIVGHH17 pKa = 6.6GLLQEE22 pKa = 4.15WSTRR26 pKa = 11.84PEE28 pKa = 4.25PPDD31 pKa = 3.72EE32 pKa = 4.47STMSDD37 pKa = 3.24PPRR40 pKa = 11.84PVKK43 pKa = 10.37ARR45 pKa = 11.84CPFRR49 pKa = 11.84LIKK52 pKa = 10.79DD53 pKa = 3.46PAITLLGKK61 pKa = 10.11FRR63 pKa = 11.84TVRR66 pKa = 11.84SSLPIDD72 pKa = 4.22PEE74 pKa = 4.8SITPGQWVSQRR85 pKa = 11.84VRR87 pKa = 11.84VTKK90 pKa = 7.2YY91 pKa = 7.39TCQWGYY97 pKa = 7.6PTIVHH102 pKa = 5.12VAVRR106 pKa = 11.84HH107 pKa = 4.8VVPSLKK113 pKa = 10.24DD114 pKa = 3.52LKK116 pKa = 11.0NLQQVNMDD124 pKa = 3.93GEE126 pKa = 4.59TQITSLPCPWYY137 pKa = 8.41PWSGTHH143 pKa = 6.56IMKK146 pKa = 10.15QEE148 pKa = 3.86SVIRR152 pKa = 11.84EE153 pKa = 3.97NKK155 pKa = 9.37SVRR158 pKa = 11.84FHH160 pKa = 6.86IGSSSYY166 pKa = 11.13FDD168 pKa = 4.03FDD170 pKa = 4.49FPLWTCKK177 pKa = 10.35LPPCVTTNPAGMWIPNGGYY196 pKa = 9.63PSQCSSWHH204 pKa = 5.83EE205 pKa = 3.98IEE207 pKa = 5.71LEE209 pKa = 3.93LLGVTRR215 pKa = 11.84ADD217 pKa = 3.34PEE219 pKa = 4.54TYY221 pKa = 9.85HH222 pKa = 6.15VVGMYY227 pKa = 9.94SYY229 pKa = 10.14PVPLRR234 pKa = 11.84KK235 pKa = 9.75SCTCTTCSRR244 pKa = 11.84NVIVLPTGDD253 pKa = 4.2LLEE256 pKa = 5.37IPLAPQISKK265 pKa = 10.26ILSSYY270 pKa = 8.53PRR272 pKa = 11.84CSISIHH278 pKa = 6.51RR279 pKa = 11.84KK280 pKa = 7.05TGLHH284 pKa = 6.33FDD286 pKa = 3.7PRR288 pKa = 11.84GLPSEE293 pKa = 4.66LLDD296 pKa = 3.45LEE298 pKa = 4.22NYY300 pKa = 8.41YY301 pKa = 10.8RR302 pKa = 11.84CTLTHH307 pKa = 6.18LTYY310 pKa = 10.44ILRR313 pKa = 11.84PPPPQLLIPLLQSDD327 pKa = 4.41SAFSLIKK334 pKa = 10.03YY335 pKa = 8.04QLNNGYY341 pKa = 10.06LSPSLSSVGVKK352 pKa = 10.56GGDD355 pKa = 3.45SQPTHH360 pKa = 5.7SQILTLHH367 pKa = 5.84IVGALLHH374 pKa = 6.11SHH376 pKa = 7.19ADD378 pKa = 3.35QNIFTRR384 pKa = 11.84YY385 pKa = 10.02DD386 pKa = 3.59YY387 pKa = 9.4NTNWSMIDD395 pKa = 3.62DD396 pKa = 5.58IIRR399 pKa = 11.84EE400 pKa = 3.94MDD402 pKa = 3.35QTVEE406 pKa = 4.36DD407 pKa = 4.44LTAQTDD413 pKa = 3.95PEE415 pKa = 4.51KK416 pKa = 10.08TQHH419 pKa = 6.37PLNRR423 pKa = 11.84QNEE426 pKa = 4.47PIFKK430 pKa = 9.88FEE432 pKa = 4.84HH433 pKa = 5.43IMYY436 pKa = 10.22LVMLISGLLITFSTFFVISKK456 pKa = 10.15KK457 pKa = 9.35EE458 pKa = 3.79QTSEE462 pKa = 3.92VLTGNNSSTSSTGKK476 pKa = 9.0RR477 pKa = 11.84RR478 pKa = 11.84KK479 pKa = 9.38VKK481 pKa = 9.34RR482 pKa = 11.84TPEE485 pKa = 3.78VWSNPSRR492 pKa = 11.84RR493 pKa = 11.84IKK495 pKa = 9.82WNCKK499 pKa = 6.45TSCSCTHH506 pKa = 7.14ASPTKK511 pKa = 10.15FMRR514 pKa = 11.84VPSRR518 pKa = 11.84SLASGKK524 pKa = 10.32SSSSS528 pKa = 3.18
MM1 pKa = 7.31RR2 pKa = 11.84TEE4 pKa = 4.47LCLSCLLLLIVGHH17 pKa = 6.6GLLQEE22 pKa = 4.15WSTRR26 pKa = 11.84PEE28 pKa = 4.25PPDD31 pKa = 3.72EE32 pKa = 4.47STMSDD37 pKa = 3.24PPRR40 pKa = 11.84PVKK43 pKa = 10.37ARR45 pKa = 11.84CPFRR49 pKa = 11.84LIKK52 pKa = 10.79DD53 pKa = 3.46PAITLLGKK61 pKa = 10.11FRR63 pKa = 11.84TVRR66 pKa = 11.84SSLPIDD72 pKa = 4.22PEE74 pKa = 4.8SITPGQWVSQRR85 pKa = 11.84VRR87 pKa = 11.84VTKK90 pKa = 7.2YY91 pKa = 7.39TCQWGYY97 pKa = 7.6PTIVHH102 pKa = 5.12VAVRR106 pKa = 11.84HH107 pKa = 4.8VVPSLKK113 pKa = 10.24DD114 pKa = 3.52LKK116 pKa = 11.0NLQQVNMDD124 pKa = 3.93GEE126 pKa = 4.59TQITSLPCPWYY137 pKa = 8.41PWSGTHH143 pKa = 6.56IMKK146 pKa = 10.15QEE148 pKa = 3.86SVIRR152 pKa = 11.84EE153 pKa = 3.97NKK155 pKa = 9.37SVRR158 pKa = 11.84FHH160 pKa = 6.86IGSSSYY166 pKa = 11.13FDD168 pKa = 4.03FDD170 pKa = 4.49FPLWTCKK177 pKa = 10.35LPPCVTTNPAGMWIPNGGYY196 pKa = 9.63PSQCSSWHH204 pKa = 5.83EE205 pKa = 3.98IEE207 pKa = 5.71LEE209 pKa = 3.93LLGVTRR215 pKa = 11.84ADD217 pKa = 3.34PEE219 pKa = 4.54TYY221 pKa = 9.85HH222 pKa = 6.15VVGMYY227 pKa = 9.94SYY229 pKa = 10.14PVPLRR234 pKa = 11.84KK235 pKa = 9.75SCTCTTCSRR244 pKa = 11.84NVIVLPTGDD253 pKa = 4.2LLEE256 pKa = 5.37IPLAPQISKK265 pKa = 10.26ILSSYY270 pKa = 8.53PRR272 pKa = 11.84CSISIHH278 pKa = 6.51RR279 pKa = 11.84KK280 pKa = 7.05TGLHH284 pKa = 6.33FDD286 pKa = 3.7PRR288 pKa = 11.84GLPSEE293 pKa = 4.66LLDD296 pKa = 3.45LEE298 pKa = 4.22NYY300 pKa = 8.41YY301 pKa = 10.8RR302 pKa = 11.84CTLTHH307 pKa = 6.18LTYY310 pKa = 10.44ILRR313 pKa = 11.84PPPPQLLIPLLQSDD327 pKa = 4.41SAFSLIKK334 pKa = 10.03YY335 pKa = 8.04QLNNGYY341 pKa = 10.06LSPSLSSVGVKK352 pKa = 10.56GGDD355 pKa = 3.45SQPTHH360 pKa = 5.7SQILTLHH367 pKa = 5.84IVGALLHH374 pKa = 6.11SHH376 pKa = 7.19ADD378 pKa = 3.35QNIFTRR384 pKa = 11.84YY385 pKa = 10.02DD386 pKa = 3.59YY387 pKa = 9.4NTNWSMIDD395 pKa = 3.62DD396 pKa = 5.58IIRR399 pKa = 11.84EE400 pKa = 3.94MDD402 pKa = 3.35QTVEE406 pKa = 4.36DD407 pKa = 4.44LTAQTDD413 pKa = 3.95PEE415 pKa = 4.51KK416 pKa = 10.08TQHH419 pKa = 6.37PLNRR423 pKa = 11.84QNEE426 pKa = 4.47PIFKK430 pKa = 9.88FEE432 pKa = 4.84HH433 pKa = 5.43IMYY436 pKa = 10.22LVMLISGLLITFSTFFVISKK456 pKa = 10.15KK457 pKa = 9.35EE458 pKa = 3.79QTSEE462 pKa = 3.92VLTGNNSSTSSTGKK476 pKa = 9.0RR477 pKa = 11.84RR478 pKa = 11.84KK479 pKa = 9.38VKK481 pKa = 9.34RR482 pKa = 11.84TPEE485 pKa = 3.78VWSNPSRR492 pKa = 11.84RR493 pKa = 11.84IKK495 pKa = 9.82WNCKK499 pKa = 6.45TSCSCTHH506 pKa = 7.14ASPTKK511 pKa = 10.15FMRR514 pKa = 11.84VPSRR518 pKa = 11.84SLASGKK524 pKa = 10.32SSSSS528 pKa = 3.18
Molecular weight: 59.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3874 |
272 |
2193 |
774.8 |
86.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.453 ± 0.976 | 1.936 ± 0.239 |
5.679 ± 0.563 | 5.395 ± 0.351 |
3.82 ± 0.66 | 5.627 ± 0.224 |
3.072 ± 0.296 | 4.75 ± 0.383 |
3.975 ± 0.458 | 10.119 ± 1.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.153 | 3.149 ± 0.333 |
6.298 ± 1.189 | 3.975 ± 0.354 |
6.531 ± 0.213 | 8.854 ± 0.683 |
6.608 ± 0.473 | 6.557 ± 0.127 |
1.91 ± 0.144 | 2.839 ± 0.25 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
