
Harrison Dam virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sunrhavirus; Harrison sunrhavirus
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
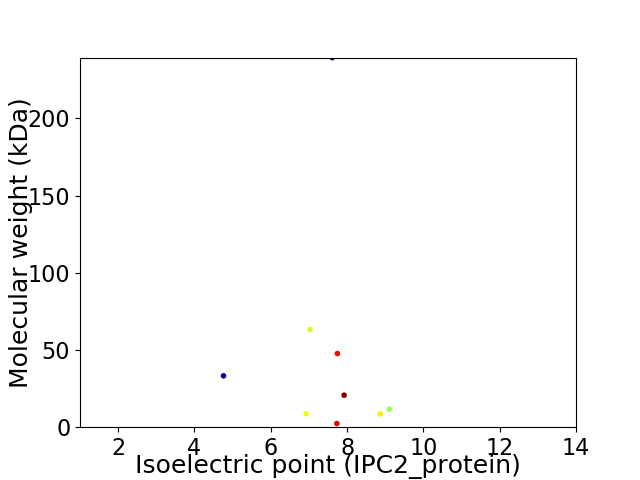
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
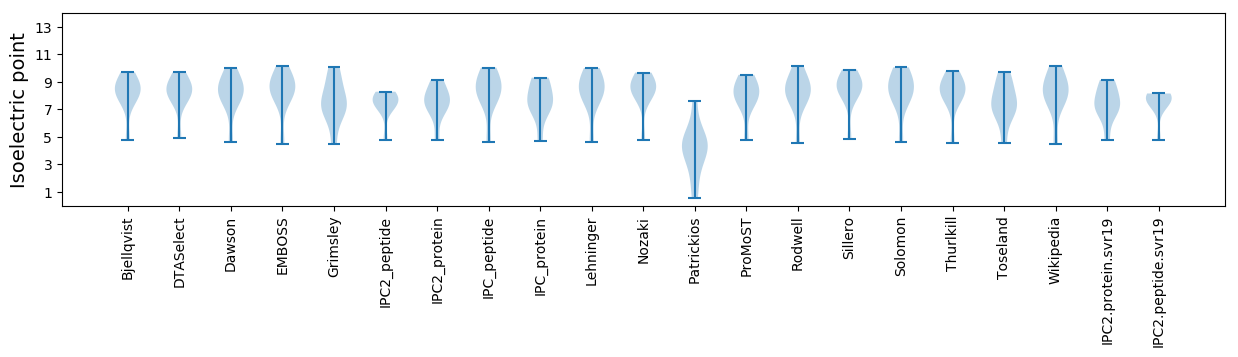
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0V1I6|A0A0A0V1I6_9RHAB Isoform of A0A0A0V5M3 C' protein OS=Harrison Dam virus OX=1569259 GN=P PE=4 SV=1
MM1 pKa = 7.49LSSKK5 pKa = 8.41TVEE8 pKa = 4.21NLTKK12 pKa = 10.62YY13 pKa = 10.3QEE15 pKa = 4.24AVQNEE20 pKa = 4.49PDD22 pKa = 4.26PSDD25 pKa = 3.87GDD27 pKa = 4.07LGDD30 pKa = 4.29SEE32 pKa = 6.13SMPQLEE38 pKa = 4.26LSGGVSRR45 pKa = 11.84EE46 pKa = 3.69SSAQLVEE53 pKa = 4.86FMQRR57 pKa = 11.84SNLADD62 pKa = 3.74GGACDD67 pKa = 5.52VPYY70 pKa = 10.1PWTADD75 pKa = 3.7SIHH78 pKa = 6.74NDD80 pKa = 2.58AFVNPEE86 pKa = 3.85GEE88 pKa = 4.17EE89 pKa = 4.04CRR91 pKa = 11.84DD92 pKa = 4.07LIDD95 pKa = 5.35EE96 pKa = 4.57SDD98 pKa = 3.67CGEE101 pKa = 4.12NSWSGRR107 pKa = 11.84RR108 pKa = 11.84SRR110 pKa = 11.84QIYY113 pKa = 8.57EE114 pKa = 3.7QGFEE118 pKa = 4.17DD119 pKa = 4.39GVRR122 pKa = 11.84EE123 pKa = 3.99VGRR126 pKa = 11.84QFRR129 pKa = 11.84MLQEE133 pKa = 3.84EE134 pKa = 4.86GFPFEE139 pKa = 4.28FDD141 pKa = 3.73YY142 pKa = 11.44VDD144 pKa = 3.76GRR146 pKa = 11.84IKK148 pKa = 10.57VKK150 pKa = 10.57KK151 pKa = 8.28ITCDD155 pKa = 3.16QRR157 pKa = 11.84ILIRR161 pKa = 11.84KK162 pKa = 8.51LIEE165 pKa = 4.31GGFFEE170 pKa = 5.6HH171 pKa = 7.34LSTDD175 pKa = 3.31MAVKK179 pKa = 10.09QKK181 pKa = 10.14SVKK184 pKa = 9.42PDD186 pKa = 3.18QPVAPLHH193 pKa = 6.13QILKK197 pKa = 8.67EE198 pKa = 4.14AKK200 pKa = 9.94KK201 pKa = 8.6PTQKK205 pKa = 10.98SMQNQEE211 pKa = 4.32EE212 pKa = 4.91KK213 pKa = 10.62FQQPLNTNSKK223 pKa = 8.83SAAMEE228 pKa = 3.92YY229 pKa = 10.39RR230 pKa = 11.84LSFCGDD236 pKa = 3.23DD237 pKa = 3.52TLTMEE242 pKa = 5.05EE243 pKa = 4.79FIEE246 pKa = 4.26KK247 pKa = 10.33ANRR250 pKa = 11.84GLHH253 pKa = 5.04ICKK256 pKa = 10.0DD257 pKa = 3.38GKK259 pKa = 10.55NRR261 pKa = 11.84FVDD264 pKa = 4.75LDD266 pKa = 3.99DD267 pKa = 4.92FCPQKK272 pKa = 10.34IRR274 pKa = 11.84EE275 pKa = 4.17RR276 pKa = 11.84KK277 pKa = 9.51ADD279 pKa = 4.01LLTLEE284 pKa = 4.29EE285 pKa = 4.61WLMCLYY291 pKa = 10.89
MM1 pKa = 7.49LSSKK5 pKa = 8.41TVEE8 pKa = 4.21NLTKK12 pKa = 10.62YY13 pKa = 10.3QEE15 pKa = 4.24AVQNEE20 pKa = 4.49PDD22 pKa = 4.26PSDD25 pKa = 3.87GDD27 pKa = 4.07LGDD30 pKa = 4.29SEE32 pKa = 6.13SMPQLEE38 pKa = 4.26LSGGVSRR45 pKa = 11.84EE46 pKa = 3.69SSAQLVEE53 pKa = 4.86FMQRR57 pKa = 11.84SNLADD62 pKa = 3.74GGACDD67 pKa = 5.52VPYY70 pKa = 10.1PWTADD75 pKa = 3.7SIHH78 pKa = 6.74NDD80 pKa = 2.58AFVNPEE86 pKa = 3.85GEE88 pKa = 4.17EE89 pKa = 4.04CRR91 pKa = 11.84DD92 pKa = 4.07LIDD95 pKa = 5.35EE96 pKa = 4.57SDD98 pKa = 3.67CGEE101 pKa = 4.12NSWSGRR107 pKa = 11.84RR108 pKa = 11.84SRR110 pKa = 11.84QIYY113 pKa = 8.57EE114 pKa = 3.7QGFEE118 pKa = 4.17DD119 pKa = 4.39GVRR122 pKa = 11.84EE123 pKa = 3.99VGRR126 pKa = 11.84QFRR129 pKa = 11.84MLQEE133 pKa = 3.84EE134 pKa = 4.86GFPFEE139 pKa = 4.28FDD141 pKa = 3.73YY142 pKa = 11.44VDD144 pKa = 3.76GRR146 pKa = 11.84IKK148 pKa = 10.57VKK150 pKa = 10.57KK151 pKa = 8.28ITCDD155 pKa = 3.16QRR157 pKa = 11.84ILIRR161 pKa = 11.84KK162 pKa = 8.51LIEE165 pKa = 4.31GGFFEE170 pKa = 5.6HH171 pKa = 7.34LSTDD175 pKa = 3.31MAVKK179 pKa = 10.09QKK181 pKa = 10.14SVKK184 pKa = 9.42PDD186 pKa = 3.18QPVAPLHH193 pKa = 6.13QILKK197 pKa = 8.67EE198 pKa = 4.14AKK200 pKa = 9.94KK201 pKa = 8.6PTQKK205 pKa = 10.98SMQNQEE211 pKa = 4.32EE212 pKa = 4.91KK213 pKa = 10.62FQQPLNTNSKK223 pKa = 8.83SAAMEE228 pKa = 3.92YY229 pKa = 10.39RR230 pKa = 11.84LSFCGDD236 pKa = 3.23DD237 pKa = 3.52TLTMEE242 pKa = 5.05EE243 pKa = 4.79FIEE246 pKa = 4.26KK247 pKa = 10.33ANRR250 pKa = 11.84GLHH253 pKa = 5.04ICKK256 pKa = 10.0DD257 pKa = 3.38GKK259 pKa = 10.55NRR261 pKa = 11.84FVDD264 pKa = 4.75LDD266 pKa = 3.99DD267 pKa = 4.92FCPQKK272 pKa = 10.34IRR274 pKa = 11.84EE275 pKa = 4.17RR276 pKa = 11.84KK277 pKa = 9.51ADD279 pKa = 4.01LLTLEE284 pKa = 4.29EE285 pKa = 4.61WLMCLYY291 pKa = 10.89
Molecular weight: 33.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0V7W9|A0A0A0V7W9_9RHAB Isoform of A0A0A0V5M3 C'' protein OS=Harrison Dam virus OX=1569259 GN=P PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84SEE4 pKa = 5.14DD5 pKa = 4.03LDD7 pKa = 4.15SEE9 pKa = 4.35INRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84ILRR17 pKa = 11.84ASIHH21 pKa = 5.2RR22 pKa = 11.84HH23 pKa = 4.58GGEE26 pKa = 3.94AEE28 pKa = 3.82ISQTRR33 pKa = 11.84STSSPVASNPEE44 pKa = 3.96RR45 pKa = 11.84SQKK48 pKa = 10.83ANPEE52 pKa = 3.8IYY54 pKa = 10.35AEE56 pKa = 4.03PRR58 pKa = 11.84GKK60 pKa = 10.62VSTTFEE66 pKa = 4.32YY67 pKa = 10.66EE68 pKa = 3.97FKK70 pKa = 10.47IGRR73 pKa = 11.84DD74 pKa = 3.87GISSKK79 pKa = 10.75LLWGRR84 pKa = 11.84HH85 pKa = 4.33INNGGIYY92 pKa = 9.72RR93 pKa = 11.84KK94 pKa = 9.7SEE96 pKa = 3.5SWTTYY101 pKa = 10.25LL102 pKa = 5.1
MM1 pKa = 7.75RR2 pKa = 11.84SEE4 pKa = 5.14DD5 pKa = 4.03LDD7 pKa = 4.15SEE9 pKa = 4.35INRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84ILRR17 pKa = 11.84ASIHH21 pKa = 5.2RR22 pKa = 11.84HH23 pKa = 4.58GGEE26 pKa = 3.94AEE28 pKa = 3.82ISQTRR33 pKa = 11.84STSSPVASNPEE44 pKa = 3.96RR45 pKa = 11.84SQKK48 pKa = 10.83ANPEE52 pKa = 3.8IYY54 pKa = 10.35AEE56 pKa = 4.03PRR58 pKa = 11.84GKK60 pKa = 10.62VSTTFEE66 pKa = 4.32YY67 pKa = 10.66EE68 pKa = 3.97FKK70 pKa = 10.47IGRR73 pKa = 11.84DD74 pKa = 3.87GISSKK79 pKa = 10.75LLWGRR84 pKa = 11.84HH85 pKa = 4.33INNGGIYY92 pKa = 9.72RR93 pKa = 11.84KK94 pKa = 9.7SEE96 pKa = 3.5SWTTYY101 pKa = 10.25LL102 pKa = 5.1
Molecular weight: 11.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3775 |
19 |
2074 |
419.4 |
48.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.841 ± 0.813 | 2.04 ± 0.177 |
5.669 ± 0.527 | 7.046 ± 0.451 |
4.795 ± 0.27 | 5.589 ± 0.49 |
2.093 ± 0.263 | 8.477 ± 0.639 |
8.185 ± 0.428 | 9.642 ± 0.544 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.216 | 5.695 ± 0.43 |
3.656 ± 0.261 | 3.576 ± 0.407 |
5.113 ± 0.29 | 7.523 ± 0.828 |
4.874 ± 0.198 | 4.371 ± 0.29 |
1.854 ± 0.25 | 3.603 ± 0.425 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
