
Torque teno virus (isolate Human/China/CT23F/2001) (TTV)
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
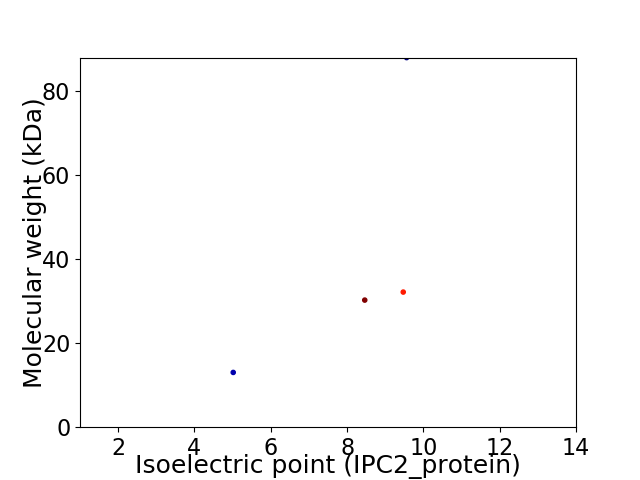
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8V7J0|CAPSD_TTVV6 Capsid protein OS=Torque teno virus (isolate Human/China/CT23F/2001) OX=687366 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.69PWRR4 pKa = 11.84PPAHH8 pKa = 5.98SVPGRR13 pKa = 11.84EE14 pKa = 4.23GQFYY18 pKa = 10.42AATFHH23 pKa = 6.52AHH25 pKa = 7.07AAFCGCGGFIEE36 pKa = 4.92HH37 pKa = 7.13LNSIHH42 pKa = 6.81PRR44 pKa = 11.84FLGAGGPPPPPPALRR59 pKa = 11.84RR60 pKa = 11.84ALPAPEE66 pKa = 5.03GPGGPPQHH74 pKa = 6.91APPNPPPEE82 pKa = 5.29GDD84 pKa = 3.39HH85 pKa = 5.59QPPRR89 pKa = 11.84RR90 pKa = 11.84GGGAGGAGDD99 pKa = 3.84GHH101 pKa = 8.29AGDD104 pKa = 4.26GDD106 pKa = 3.72AAEE109 pKa = 4.64EE110 pKa = 4.15YY111 pKa = 10.97GPEE114 pKa = 4.51DD115 pKa = 4.92LDD117 pKa = 4.72LLFAAAAEE125 pKa = 4.26DD126 pKa = 4.55DD127 pKa = 3.84MM128 pKa = 7.94
MM1 pKa = 7.69PWRR4 pKa = 11.84PPAHH8 pKa = 5.98SVPGRR13 pKa = 11.84EE14 pKa = 4.23GQFYY18 pKa = 10.42AATFHH23 pKa = 6.52AHH25 pKa = 7.07AAFCGCGGFIEE36 pKa = 4.92HH37 pKa = 7.13LNSIHH42 pKa = 6.81PRR44 pKa = 11.84FLGAGGPPPPPPALRR59 pKa = 11.84RR60 pKa = 11.84ALPAPEE66 pKa = 5.03GPGGPPQHH74 pKa = 6.91APPNPPPEE82 pKa = 5.29GDD84 pKa = 3.39HH85 pKa = 5.59QPPRR89 pKa = 11.84RR90 pKa = 11.84GGGAGGAGDD99 pKa = 3.84GHH101 pKa = 8.29AGDD104 pKa = 4.26GDD106 pKa = 3.72AAEE109 pKa = 4.64EE110 pKa = 4.15YY111 pKa = 10.97GPEE114 pKa = 4.51DD115 pKa = 4.92LDD117 pKa = 4.72LLFAAAAEE125 pKa = 4.26DD126 pKa = 4.55DD127 pKa = 3.84MM128 pKa = 7.94
Molecular weight: 13.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8V7J2|ORF24_TTVV6 Uncharacterized ORF2/4 protein OS=Torque teno virus (isolate Human/China/CT23F/2001) OX=687366 GN=ORF2/4 PE=4 SV=1
MM1 pKa = 7.69PWRR4 pKa = 11.84PPAHH8 pKa = 5.98SVPGRR13 pKa = 11.84EE14 pKa = 4.23GQFYY18 pKa = 10.42AATFHH23 pKa = 6.52AHH25 pKa = 7.07AAFCGCGGFIEE36 pKa = 4.92HH37 pKa = 7.13LNSIHH42 pKa = 6.81PRR44 pKa = 11.84FLGAGGPPPPPPALRR59 pKa = 11.84RR60 pKa = 11.84ALPAPEE66 pKa = 5.03GPGGPPQHH74 pKa = 6.91APPNPPPEE82 pKa = 5.29GDD84 pKa = 3.39HH85 pKa = 5.59QPPRR89 pKa = 11.84RR90 pKa = 11.84GGGAGGAGDD99 pKa = 3.84GHH101 pKa = 8.29AGDD104 pKa = 4.26GDD106 pKa = 3.72AAEE109 pKa = 4.64EE110 pKa = 4.15YY111 pKa = 10.97GPEE114 pKa = 4.51DD115 pKa = 4.92LDD117 pKa = 4.72LLFAAAAEE125 pKa = 4.43DD126 pKa = 4.04DD127 pKa = 4.18MSFKK131 pKa = 10.92APSSRR136 pKa = 11.84HH137 pKa = 3.71QTRR140 pKa = 11.84GPGRR144 pKa = 11.84RR145 pKa = 11.84AKK147 pKa = 10.26KK148 pKa = 9.96RR149 pKa = 11.84LRR151 pKa = 11.84FSPGSPRR158 pKa = 11.84QPRR161 pKa = 11.84LGGEE165 pKa = 4.38SRR167 pKa = 11.84RR168 pKa = 11.84SPSPRR173 pKa = 11.84RR174 pKa = 11.84TSTPKK179 pKa = 10.0RR180 pKa = 11.84KK181 pKa = 9.8RR182 pKa = 11.84GATPQAAPAAKK193 pKa = 6.9TTPASPQTRR202 pKa = 11.84TPSPVRR208 pKa = 11.84RR209 pKa = 11.84RR210 pKa = 11.84TQTEE214 pKa = 3.74EE215 pKa = 4.3GSPHH219 pKa = 6.97RR220 pKa = 11.84PPPYY224 pKa = 9.3IAPPPIVEE232 pKa = 5.29DD233 pKa = 3.62LLFPNTQKK241 pKa = 10.86KK242 pKa = 10.29KK243 pKa = 10.52KK244 pKa = 9.61FSKK247 pKa = 10.52FDD249 pKa = 3.46WEE251 pKa = 4.46TEE253 pKa = 4.0AQLAACFDD261 pKa = 3.52RR262 pKa = 11.84PMRR265 pKa = 11.84FFPSDD270 pKa = 3.37LPTYY274 pKa = 9.5PWLPKK279 pKa = 10.41KK280 pKa = 9.29PTTQTTFRR288 pKa = 11.84VSFQLKK294 pKa = 10.05APQQ297 pKa = 3.2
MM1 pKa = 7.69PWRR4 pKa = 11.84PPAHH8 pKa = 5.98SVPGRR13 pKa = 11.84EE14 pKa = 4.23GQFYY18 pKa = 10.42AATFHH23 pKa = 6.52AHH25 pKa = 7.07AAFCGCGGFIEE36 pKa = 4.92HH37 pKa = 7.13LNSIHH42 pKa = 6.81PRR44 pKa = 11.84FLGAGGPPPPPPALRR59 pKa = 11.84RR60 pKa = 11.84ALPAPEE66 pKa = 5.03GPGGPPQHH74 pKa = 6.91APPNPPPEE82 pKa = 5.29GDD84 pKa = 3.39HH85 pKa = 5.59QPPRR89 pKa = 11.84RR90 pKa = 11.84GGGAGGAGDD99 pKa = 3.84GHH101 pKa = 8.29AGDD104 pKa = 4.26GDD106 pKa = 3.72AAEE109 pKa = 4.64EE110 pKa = 4.15YY111 pKa = 10.97GPEE114 pKa = 4.51DD115 pKa = 4.92LDD117 pKa = 4.72LLFAAAAEE125 pKa = 4.43DD126 pKa = 4.04DD127 pKa = 4.18MSFKK131 pKa = 10.92APSSRR136 pKa = 11.84HH137 pKa = 3.71QTRR140 pKa = 11.84GPGRR144 pKa = 11.84RR145 pKa = 11.84AKK147 pKa = 10.26KK148 pKa = 9.96RR149 pKa = 11.84LRR151 pKa = 11.84FSPGSPRR158 pKa = 11.84QPRR161 pKa = 11.84LGGEE165 pKa = 4.38SRR167 pKa = 11.84RR168 pKa = 11.84SPSPRR173 pKa = 11.84RR174 pKa = 11.84TSTPKK179 pKa = 10.0RR180 pKa = 11.84KK181 pKa = 9.8RR182 pKa = 11.84GATPQAAPAAKK193 pKa = 6.9TTPASPQTRR202 pKa = 11.84TPSPVRR208 pKa = 11.84RR209 pKa = 11.84RR210 pKa = 11.84TQTEE214 pKa = 3.74EE215 pKa = 4.3GSPHH219 pKa = 6.97RR220 pKa = 11.84PPPYY224 pKa = 9.3IAPPPIVEE232 pKa = 5.29DD233 pKa = 3.62LLFPNTQKK241 pKa = 10.86KK242 pKa = 10.29KK243 pKa = 10.52KK244 pKa = 9.61FSKK247 pKa = 10.52FDD249 pKa = 3.46WEE251 pKa = 4.46TEE253 pKa = 4.0AQLAACFDD261 pKa = 3.52RR262 pKa = 11.84PMRR265 pKa = 11.84FFPSDD270 pKa = 3.37LPTYY274 pKa = 9.5PWLPKK279 pKa = 10.41KK280 pKa = 9.29PTTQTTFRR288 pKa = 11.84VSFQLKK294 pKa = 10.05APQQ297 pKa = 3.2
Molecular weight: 32.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1450 |
128 |
745 |
362.5 |
40.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.448 ± 2.001 | 1.448 ± 0.128 |
4.552 ± 0.26 | 4.897 ± 0.457 |
4.621 ± 0.329 | 8.069 ± 1.549 |
3.172 ± 0.477 | 2.69 ± 0.852 |
5.379 ± 0.724 | 7.034 ± 0.951 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.724 ± 0.269 | 2.207 ± 0.346 |
11.448 ± 2.45 | 4.414 ± 0.45 |
9.034 ± 0.81 | 6.966 ± 1.283 |
6.276 ± 0.875 | 3.034 ± 0.745 |
2.138 ± 0.765 | 3.448 ± 1.001 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
