
Pteropus alecto (Black flying fox)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Chiroptera; Megachiroptera;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
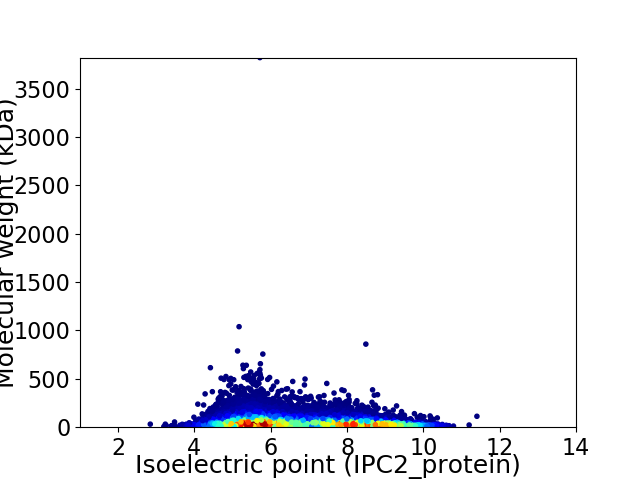
Virtual 2D-PAGE plot for 19521 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L5KQ53|L5KQ53_PTEAL Uncharacterized protein OS=Pteropus alecto OX=9402 GN=PAL_GLEAN10001291 PE=4 SV=1
MM1 pKa = 8.24DD2 pKa = 4.75FQQLADD8 pKa = 4.22VAEE11 pKa = 4.85KK12 pKa = 9.68WCSHH16 pKa = 5.29TPFEE20 pKa = 5.61LIATEE25 pKa = 3.96EE26 pKa = 4.32TEE28 pKa = 4.33RR29 pKa = 11.84RR30 pKa = 11.84MDD32 pKa = 4.29FYY34 pKa = 11.47ADD36 pKa = 3.74PGVSFYY42 pKa = 10.98VLCPDD47 pKa = 4.64NGCGDD52 pKa = 3.96NFHH55 pKa = 6.3VWSEE59 pKa = 4.48SEE61 pKa = 3.97DD62 pKa = 3.63CLPFLQLAQDD72 pKa = 4.45YY73 pKa = 10.71ISSCGKK79 pKa = 8.66KK80 pKa = 8.01TLQEE84 pKa = 3.92VLEE87 pKa = 4.35KK88 pKa = 10.64VFKK91 pKa = 10.63SFRR94 pKa = 11.84PLLGLPDD101 pKa = 4.91ADD103 pKa = 4.57DD104 pKa = 4.81DD105 pKa = 4.54ALEE108 pKa = 4.66EE109 pKa = 4.06YY110 pKa = 10.6SADD113 pKa = 3.71VEE115 pKa = 4.38EE116 pKa = 5.09EE117 pKa = 4.2EE118 pKa = 5.29PEE120 pKa = 4.13ADD122 pKa = 3.66HH123 pKa = 6.65PQMGVSQQQ131 pKa = 3.0
MM1 pKa = 8.24DD2 pKa = 4.75FQQLADD8 pKa = 4.22VAEE11 pKa = 4.85KK12 pKa = 9.68WCSHH16 pKa = 5.29TPFEE20 pKa = 5.61LIATEE25 pKa = 3.96EE26 pKa = 4.32TEE28 pKa = 4.33RR29 pKa = 11.84RR30 pKa = 11.84MDD32 pKa = 4.29FYY34 pKa = 11.47ADD36 pKa = 3.74PGVSFYY42 pKa = 10.98VLCPDD47 pKa = 4.64NGCGDD52 pKa = 3.96NFHH55 pKa = 6.3VWSEE59 pKa = 4.48SEE61 pKa = 3.97DD62 pKa = 3.63CLPFLQLAQDD72 pKa = 4.45YY73 pKa = 10.71ISSCGKK79 pKa = 8.66KK80 pKa = 8.01TLQEE84 pKa = 3.92VLEE87 pKa = 4.35KK88 pKa = 10.64VFKK91 pKa = 10.63SFRR94 pKa = 11.84PLLGLPDD101 pKa = 4.91ADD103 pKa = 4.57DD104 pKa = 4.81DD105 pKa = 4.54ALEE108 pKa = 4.66EE109 pKa = 4.06YY110 pKa = 10.6SADD113 pKa = 3.71VEE115 pKa = 4.38EE116 pKa = 5.09EE117 pKa = 4.2EE118 pKa = 5.29PEE120 pKa = 4.13ADD122 pKa = 3.66HH123 pKa = 6.65PQMGVSQQQ131 pKa = 3.0
Molecular weight: 14.91 kDa
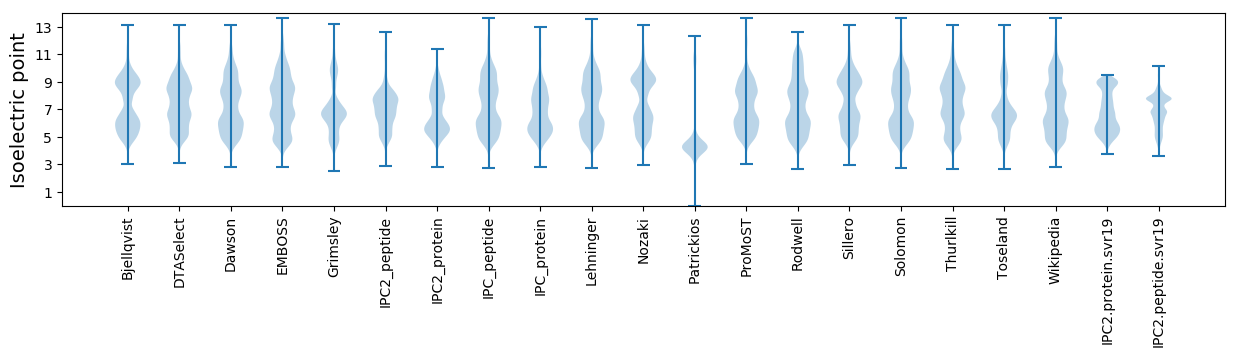
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L5KK91|L5KK91_PTEAL CDP-diacylglycerol--inositol 3-phosphatidyltransferase OS=Pteropus alecto OX=9402 GN=PAL_GLEAN10011874 PE=3 SV=1
MM1 pKa = 7.47PHH3 pKa = 5.92QPRR6 pKa = 11.84HH7 pKa = 5.17RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84SSHH15 pKa = 5.15SQAGHH20 pKa = 6.02SGTRR24 pKa = 11.84SPPAPRR30 pKa = 11.84SPVPPRR36 pKa = 11.84SPVPPRR42 pKa = 11.84SPVPPRR48 pKa = 11.84SPVPPRR54 pKa = 11.84SPVPPRR60 pKa = 11.84SPVPPRR66 pKa = 11.84SPVPPRR72 pKa = 11.84SPVPPRR78 pKa = 11.84SPVPPRR84 pKa = 11.84SPVPPRR90 pKa = 11.84SPVPPRR96 pKa = 11.84SPVPPRR102 pKa = 11.84SPVPPRR108 pKa = 11.84SPVPPRR114 pKa = 11.84SPVPPRR120 pKa = 11.84SPVPPRR126 pKa = 11.84SPVPPRR132 pKa = 11.84SPVPPRR138 pKa = 11.84SPVPPRR144 pKa = 11.84SPVPPRR150 pKa = 11.84SPVPPRR156 pKa = 11.84SPVPPRR162 pKa = 11.84SPVPPRR168 pKa = 11.84SPVPPRR174 pKa = 11.84SPVPPRR180 pKa = 11.84SPVPPRR186 pKa = 11.84SPVPPRR192 pKa = 11.84SPVPPRR198 pKa = 11.84SPVPPRR204 pKa = 11.84SPVPPRR210 pKa = 11.84SPVPPRR216 pKa = 11.84SPVPPRR222 pKa = 11.84SPVPPRR228 pKa = 11.84SPVPPRR234 pKa = 11.84SPVPPRR240 pKa = 11.84SPVPPRR246 pKa = 11.84SPVPPRR252 pKa = 11.84SPVPPRR258 pKa = 11.84SPVPPRR264 pKa = 11.84SPVPPRR270 pKa = 11.84SPVPPRR276 pKa = 11.84SPVPPRR282 pKa = 11.84SPVPPRR288 pKa = 11.84SPVPPRR294 pKa = 11.84SPVPPRR300 pKa = 11.84SPVPPRR306 pKa = 11.84SPVPPRR312 pKa = 11.84SPVPPRR318 pKa = 11.84SPVPPRR324 pKa = 11.84SPVPPRR330 pKa = 11.84SPVPPRR336 pKa = 11.84SPVPPRR342 pKa = 11.84SPVPPRR348 pKa = 11.84SPVPPRR354 pKa = 11.84SPVPPRR360 pKa = 11.84SPVPPRR366 pKa = 11.84SPVPPRR372 pKa = 11.84SPVPPRR378 pKa = 11.84SPVPPRR384 pKa = 11.84SPVPPRR390 pKa = 11.84SPVPPRR396 pKa = 11.84SPVPPRR402 pKa = 11.84SPVPPRR408 pKa = 11.84SPVPPRR414 pKa = 11.84SPVPPRR420 pKa = 11.84SPVPPRR426 pKa = 11.84SPVPPRR432 pKa = 11.84SPVPPRR438 pKa = 11.84SPVPPRR444 pKa = 11.84SPVPPRR450 pKa = 11.84SPVPPRR456 pKa = 11.84SPVPPRR462 pKa = 11.84SPVPPRR468 pKa = 11.84SPVPPRR474 pKa = 11.84SPVPPRR480 pKa = 11.84SPVPPRR486 pKa = 11.84SPVPPRR492 pKa = 11.84SPVPPRR498 pKa = 11.84SPVPPRR504 pKa = 11.84SPVPPRR510 pKa = 11.84SPVPPRR516 pKa = 11.84SPVPPRR522 pKa = 11.84SPVPPRR528 pKa = 11.84SPVPPRR534 pKa = 11.84SPVPPRR540 pKa = 11.84SPVPPRR546 pKa = 11.84SPVPPRR552 pKa = 11.84SPVPPRR558 pKa = 11.84SPVPPRR564 pKa = 11.84SPVPPRR570 pKa = 11.84SPVPPRR576 pKa = 11.84SPVPPRR582 pKa = 11.84SPVPPRR588 pKa = 11.84SPVPPRR594 pKa = 11.84SPVPPRR600 pKa = 11.84SPVPPRR606 pKa = 11.84SPVPPRR612 pKa = 11.84SPVPPRR618 pKa = 11.84SPVPPRR624 pKa = 11.84SPVPPRR630 pKa = 11.84SPVPPRR636 pKa = 11.84SPVPPRR642 pKa = 11.84SPVPPRR648 pKa = 11.84SPVPPRR654 pKa = 11.84SPVPPRR660 pKa = 11.84SPVPPRR666 pKa = 11.84SPVPPRR672 pKa = 11.84SPVPPRR678 pKa = 11.84SPVPPRR684 pKa = 11.84SPVPPRR690 pKa = 11.84SPVPPRR696 pKa = 11.84SPVPPRR702 pKa = 11.84SPVPPRR708 pKa = 11.84SPVPPRR714 pKa = 11.84SPVPPRR720 pKa = 11.84SPVPPRR726 pKa = 11.84SPVPPRR732 pKa = 11.84SPVPPRR738 pKa = 11.84SPVPPRR744 pKa = 11.84SPVPPRR750 pKa = 11.84SPVPPRR756 pKa = 11.84SPVPPRR762 pKa = 11.84SPVPPRR768 pKa = 11.84SPVPPRR774 pKa = 11.84SPVPPRR780 pKa = 11.84SPVPPRR786 pKa = 11.84SPVPPRR792 pKa = 11.84SPVPPRR798 pKa = 11.84SPVPPRR804 pKa = 11.84SPVPPRR810 pKa = 11.84SPVPPRR816 pKa = 11.84SPVPPRR822 pKa = 11.84SPVPPRR828 pKa = 11.84SPVPPRR834 pKa = 11.84SPVPPRR840 pKa = 11.84SPVPPRR846 pKa = 11.84SPVPPRR852 pKa = 11.84SPVPPRR858 pKa = 11.84SPVPPRR864 pKa = 11.84SPVPPRR870 pKa = 11.84SPVPPRR876 pKa = 11.84SPVPPRR882 pKa = 11.84SPVPPRR888 pKa = 11.84SPVPPRR894 pKa = 11.84SPVPPRR900 pKa = 11.84SPVPPRR906 pKa = 11.84SPVPPRR912 pKa = 11.84SPVPPRR918 pKa = 11.84SPVPPRR924 pKa = 11.84SPVPPRR930 pKa = 11.84SPVPPRR936 pKa = 11.84SPVPPRR942 pKa = 11.84SPVPPRR948 pKa = 11.84SPVPPRR954 pKa = 11.84SPVPPRR960 pKa = 11.84SPVPPRR966 pKa = 11.84SPVPPRR972 pKa = 11.84SPVPPRR978 pKa = 11.84SPPAPRR984 pKa = 11.84SPPPHH989 pKa = 6.94WPTLGLLSSPVSLQGLCAEE1008 pKa = 4.44PSEE1011 pKa = 4.78VATSHH1016 pKa = 6.85LPPPSLEE1023 pKa = 3.92WQGVVGTPFSLRR1035 pKa = 11.84QTHH1038 pKa = 6.15YY1039 pKa = 10.16EE1040 pKa = 3.96LQALSSPSEE1049 pKa = 4.02LCDD1052 pKa = 3.54SGEE1055 pKa = 4.18SLNLSAPAASAVKK1068 pKa = 10.35
MM1 pKa = 7.47PHH3 pKa = 5.92QPRR6 pKa = 11.84HH7 pKa = 5.17RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84SSHH15 pKa = 5.15SQAGHH20 pKa = 6.02SGTRR24 pKa = 11.84SPPAPRR30 pKa = 11.84SPVPPRR36 pKa = 11.84SPVPPRR42 pKa = 11.84SPVPPRR48 pKa = 11.84SPVPPRR54 pKa = 11.84SPVPPRR60 pKa = 11.84SPVPPRR66 pKa = 11.84SPVPPRR72 pKa = 11.84SPVPPRR78 pKa = 11.84SPVPPRR84 pKa = 11.84SPVPPRR90 pKa = 11.84SPVPPRR96 pKa = 11.84SPVPPRR102 pKa = 11.84SPVPPRR108 pKa = 11.84SPVPPRR114 pKa = 11.84SPVPPRR120 pKa = 11.84SPVPPRR126 pKa = 11.84SPVPPRR132 pKa = 11.84SPVPPRR138 pKa = 11.84SPVPPRR144 pKa = 11.84SPVPPRR150 pKa = 11.84SPVPPRR156 pKa = 11.84SPVPPRR162 pKa = 11.84SPVPPRR168 pKa = 11.84SPVPPRR174 pKa = 11.84SPVPPRR180 pKa = 11.84SPVPPRR186 pKa = 11.84SPVPPRR192 pKa = 11.84SPVPPRR198 pKa = 11.84SPVPPRR204 pKa = 11.84SPVPPRR210 pKa = 11.84SPVPPRR216 pKa = 11.84SPVPPRR222 pKa = 11.84SPVPPRR228 pKa = 11.84SPVPPRR234 pKa = 11.84SPVPPRR240 pKa = 11.84SPVPPRR246 pKa = 11.84SPVPPRR252 pKa = 11.84SPVPPRR258 pKa = 11.84SPVPPRR264 pKa = 11.84SPVPPRR270 pKa = 11.84SPVPPRR276 pKa = 11.84SPVPPRR282 pKa = 11.84SPVPPRR288 pKa = 11.84SPVPPRR294 pKa = 11.84SPVPPRR300 pKa = 11.84SPVPPRR306 pKa = 11.84SPVPPRR312 pKa = 11.84SPVPPRR318 pKa = 11.84SPVPPRR324 pKa = 11.84SPVPPRR330 pKa = 11.84SPVPPRR336 pKa = 11.84SPVPPRR342 pKa = 11.84SPVPPRR348 pKa = 11.84SPVPPRR354 pKa = 11.84SPVPPRR360 pKa = 11.84SPVPPRR366 pKa = 11.84SPVPPRR372 pKa = 11.84SPVPPRR378 pKa = 11.84SPVPPRR384 pKa = 11.84SPVPPRR390 pKa = 11.84SPVPPRR396 pKa = 11.84SPVPPRR402 pKa = 11.84SPVPPRR408 pKa = 11.84SPVPPRR414 pKa = 11.84SPVPPRR420 pKa = 11.84SPVPPRR426 pKa = 11.84SPVPPRR432 pKa = 11.84SPVPPRR438 pKa = 11.84SPVPPRR444 pKa = 11.84SPVPPRR450 pKa = 11.84SPVPPRR456 pKa = 11.84SPVPPRR462 pKa = 11.84SPVPPRR468 pKa = 11.84SPVPPRR474 pKa = 11.84SPVPPRR480 pKa = 11.84SPVPPRR486 pKa = 11.84SPVPPRR492 pKa = 11.84SPVPPRR498 pKa = 11.84SPVPPRR504 pKa = 11.84SPVPPRR510 pKa = 11.84SPVPPRR516 pKa = 11.84SPVPPRR522 pKa = 11.84SPVPPRR528 pKa = 11.84SPVPPRR534 pKa = 11.84SPVPPRR540 pKa = 11.84SPVPPRR546 pKa = 11.84SPVPPRR552 pKa = 11.84SPVPPRR558 pKa = 11.84SPVPPRR564 pKa = 11.84SPVPPRR570 pKa = 11.84SPVPPRR576 pKa = 11.84SPVPPRR582 pKa = 11.84SPVPPRR588 pKa = 11.84SPVPPRR594 pKa = 11.84SPVPPRR600 pKa = 11.84SPVPPRR606 pKa = 11.84SPVPPRR612 pKa = 11.84SPVPPRR618 pKa = 11.84SPVPPRR624 pKa = 11.84SPVPPRR630 pKa = 11.84SPVPPRR636 pKa = 11.84SPVPPRR642 pKa = 11.84SPVPPRR648 pKa = 11.84SPVPPRR654 pKa = 11.84SPVPPRR660 pKa = 11.84SPVPPRR666 pKa = 11.84SPVPPRR672 pKa = 11.84SPVPPRR678 pKa = 11.84SPVPPRR684 pKa = 11.84SPVPPRR690 pKa = 11.84SPVPPRR696 pKa = 11.84SPVPPRR702 pKa = 11.84SPVPPRR708 pKa = 11.84SPVPPRR714 pKa = 11.84SPVPPRR720 pKa = 11.84SPVPPRR726 pKa = 11.84SPVPPRR732 pKa = 11.84SPVPPRR738 pKa = 11.84SPVPPRR744 pKa = 11.84SPVPPRR750 pKa = 11.84SPVPPRR756 pKa = 11.84SPVPPRR762 pKa = 11.84SPVPPRR768 pKa = 11.84SPVPPRR774 pKa = 11.84SPVPPRR780 pKa = 11.84SPVPPRR786 pKa = 11.84SPVPPRR792 pKa = 11.84SPVPPRR798 pKa = 11.84SPVPPRR804 pKa = 11.84SPVPPRR810 pKa = 11.84SPVPPRR816 pKa = 11.84SPVPPRR822 pKa = 11.84SPVPPRR828 pKa = 11.84SPVPPRR834 pKa = 11.84SPVPPRR840 pKa = 11.84SPVPPRR846 pKa = 11.84SPVPPRR852 pKa = 11.84SPVPPRR858 pKa = 11.84SPVPPRR864 pKa = 11.84SPVPPRR870 pKa = 11.84SPVPPRR876 pKa = 11.84SPVPPRR882 pKa = 11.84SPVPPRR888 pKa = 11.84SPVPPRR894 pKa = 11.84SPVPPRR900 pKa = 11.84SPVPPRR906 pKa = 11.84SPVPPRR912 pKa = 11.84SPVPPRR918 pKa = 11.84SPVPPRR924 pKa = 11.84SPVPPRR930 pKa = 11.84SPVPPRR936 pKa = 11.84SPVPPRR942 pKa = 11.84SPVPPRR948 pKa = 11.84SPVPPRR954 pKa = 11.84SPVPPRR960 pKa = 11.84SPVPPRR966 pKa = 11.84SPVPPRR972 pKa = 11.84SPVPPRR978 pKa = 11.84SPPAPRR984 pKa = 11.84SPPPHH989 pKa = 6.94WPTLGLLSSPVSLQGLCAEE1008 pKa = 4.44PSEE1011 pKa = 4.78VATSHH1016 pKa = 6.85LPPPSLEE1023 pKa = 3.92WQGVVGTPFSLRR1035 pKa = 11.84QTHH1038 pKa = 6.15YY1039 pKa = 10.16EE1040 pKa = 3.96LQALSSPSEE1049 pKa = 4.02LCDD1052 pKa = 3.54SGEE1055 pKa = 4.18SLNLSAPAASAVKK1068 pKa = 10.35
Molecular weight: 112.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9971383 |
28 |
34674 |
510.8 |
56.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.103 ± 0.02 | 2.213 ± 0.013 |
4.805 ± 0.012 | 7.05 ± 0.023 |
3.624 ± 0.014 | 6.536 ± 0.023 |
2.597 ± 0.009 | 4.28 ± 0.016 |
5.618 ± 0.021 | 10.033 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.009 | 3.517 ± 0.013 |
6.285 ± 0.027 | 4.726 ± 0.019 |
5.755 ± 0.017 | 8.273 ± 0.023 |
5.271 ± 0.023 | 6.106 ± 0.018 |
1.215 ± 0.006 | 2.608 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
