
Sewage-associated circular DNA virus-4
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
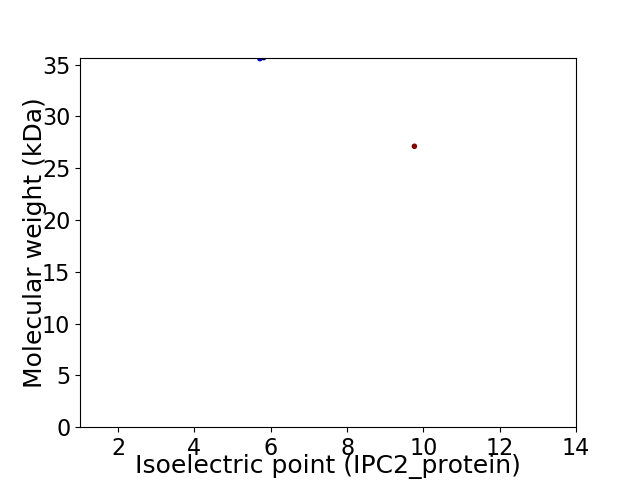
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J462|A0A075J462_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-4 OX=1519393 PE=3 SV=1
MM1 pKa = 7.48ARR3 pKa = 11.84FHH5 pKa = 7.0AKK7 pKa = 8.6TVFLTYY13 pKa = 9.06PQCPVNKK20 pKa = 9.38EE21 pKa = 4.19DD22 pKa = 4.8LLAWLQTLRR31 pKa = 11.84NPIYY35 pKa = 10.11TIVSHH40 pKa = 6.13EE41 pKa = 4.03LHH43 pKa = 6.97EE44 pKa = 5.46DD45 pKa = 4.13GSPHH49 pKa = 5.93LHH51 pKa = 7.02AVVCWQNKK59 pKa = 9.12LDD61 pKa = 4.01IRR63 pKa = 11.84DD64 pKa = 3.73TTFFNYY70 pKa = 9.84EE71 pKa = 3.93NYY73 pKa = 10.13HH74 pKa = 6.41PNIQSARR81 pKa = 11.84SIKK84 pKa = 10.41KK85 pKa = 9.74VIAYY89 pKa = 6.93TKK91 pKa = 10.44KK92 pKa = 10.6DD93 pKa = 3.06GDD95 pKa = 4.02FIEE98 pKa = 6.23AGDD101 pKa = 4.07SPIVDD106 pKa = 4.08TANTWSTAMDD116 pKa = 3.53TATTIKK122 pKa = 10.8EE123 pKa = 3.97FMDD126 pKa = 3.68TVKK129 pKa = 10.65EE130 pKa = 4.32SNPRR134 pKa = 11.84DD135 pKa = 3.6YY136 pKa = 10.92ILQHH140 pKa = 5.82EE141 pKa = 4.27RR142 pKa = 11.84LEE144 pKa = 4.32YY145 pKa = 10.19FAAKK149 pKa = 9.9HH150 pKa = 5.24FKK152 pKa = 10.46KK153 pKa = 9.96DD154 pKa = 3.39TEE156 pKa = 4.44VYY158 pKa = 10.08KK159 pKa = 11.0PNFEE163 pKa = 4.25EE164 pKa = 5.58FILPHH169 pKa = 6.32EE170 pKa = 4.46VEE172 pKa = 3.63AWLNEE177 pKa = 3.84EE178 pKa = 4.21FNSDD182 pKa = 3.62KK183 pKa = 10.85LRR185 pKa = 11.84KK186 pKa = 9.35KK187 pKa = 10.56SLILIGPSKK196 pKa = 10.5LGKK199 pKa = 8.13TEE201 pKa = 3.82WARR204 pKa = 11.84SLGPHH209 pKa = 6.44MYY211 pKa = 10.19FNHH214 pKa = 6.89LANFKK219 pKa = 10.76DD220 pKa = 3.77DD221 pKa = 3.89WDD223 pKa = 5.2DD224 pKa = 3.35EE225 pKa = 4.4AKK227 pKa = 11.1YY228 pKa = 10.58IVFDD232 pKa = 4.84DD233 pKa = 4.08FDD235 pKa = 6.0FDD237 pKa = 5.53FIPNKK242 pKa = 10.23KK243 pKa = 10.25GFFGGQQHH251 pKa = 5.61FQITGKK257 pKa = 9.21YY258 pKa = 7.38MRR260 pKa = 11.84VKK262 pKa = 9.64SVRR265 pKa = 11.84WGKK268 pKa = 10.21CCIYY272 pKa = 9.98CANVTPNIKK281 pKa = 10.62DD282 pKa = 3.41IDD284 pKa = 4.02KK285 pKa = 10.8DD286 pKa = 3.61WYY288 pKa = 11.06NEE290 pKa = 3.6NCVFIDD296 pKa = 3.75VVNKK300 pKa = 9.45FYY302 pKa = 11.39
MM1 pKa = 7.48ARR3 pKa = 11.84FHH5 pKa = 7.0AKK7 pKa = 8.6TVFLTYY13 pKa = 9.06PQCPVNKK20 pKa = 9.38EE21 pKa = 4.19DD22 pKa = 4.8LLAWLQTLRR31 pKa = 11.84NPIYY35 pKa = 10.11TIVSHH40 pKa = 6.13EE41 pKa = 4.03LHH43 pKa = 6.97EE44 pKa = 5.46DD45 pKa = 4.13GSPHH49 pKa = 5.93LHH51 pKa = 7.02AVVCWQNKK59 pKa = 9.12LDD61 pKa = 4.01IRR63 pKa = 11.84DD64 pKa = 3.73TTFFNYY70 pKa = 9.84EE71 pKa = 3.93NYY73 pKa = 10.13HH74 pKa = 6.41PNIQSARR81 pKa = 11.84SIKK84 pKa = 10.41KK85 pKa = 9.74VIAYY89 pKa = 6.93TKK91 pKa = 10.44KK92 pKa = 10.6DD93 pKa = 3.06GDD95 pKa = 4.02FIEE98 pKa = 6.23AGDD101 pKa = 4.07SPIVDD106 pKa = 4.08TANTWSTAMDD116 pKa = 3.53TATTIKK122 pKa = 10.8EE123 pKa = 3.97FMDD126 pKa = 3.68TVKK129 pKa = 10.65EE130 pKa = 4.32SNPRR134 pKa = 11.84DD135 pKa = 3.6YY136 pKa = 10.92ILQHH140 pKa = 5.82EE141 pKa = 4.27RR142 pKa = 11.84LEE144 pKa = 4.32YY145 pKa = 10.19FAAKK149 pKa = 9.9HH150 pKa = 5.24FKK152 pKa = 10.46KK153 pKa = 9.96DD154 pKa = 3.39TEE156 pKa = 4.44VYY158 pKa = 10.08KK159 pKa = 11.0PNFEE163 pKa = 4.25EE164 pKa = 5.58FILPHH169 pKa = 6.32EE170 pKa = 4.46VEE172 pKa = 3.63AWLNEE177 pKa = 3.84EE178 pKa = 4.21FNSDD182 pKa = 3.62KK183 pKa = 10.85LRR185 pKa = 11.84KK186 pKa = 9.35KK187 pKa = 10.56SLILIGPSKK196 pKa = 10.5LGKK199 pKa = 8.13TEE201 pKa = 3.82WARR204 pKa = 11.84SLGPHH209 pKa = 6.44MYY211 pKa = 10.19FNHH214 pKa = 6.89LANFKK219 pKa = 10.76DD220 pKa = 3.77DD221 pKa = 3.89WDD223 pKa = 5.2DD224 pKa = 3.35EE225 pKa = 4.4AKK227 pKa = 11.1YY228 pKa = 10.58IVFDD232 pKa = 4.84DD233 pKa = 4.08FDD235 pKa = 6.0FDD237 pKa = 5.53FIPNKK242 pKa = 10.23KK243 pKa = 10.25GFFGGQQHH251 pKa = 5.61FQITGKK257 pKa = 9.21YY258 pKa = 7.38MRR260 pKa = 11.84VKK262 pKa = 9.64SVRR265 pKa = 11.84WGKK268 pKa = 10.21CCIYY272 pKa = 9.98CANVTPNIKK281 pKa = 10.62DD282 pKa = 3.41IDD284 pKa = 4.02KK285 pKa = 10.8DD286 pKa = 3.61WYY288 pKa = 11.06NEE290 pKa = 3.6NCVFIDD296 pKa = 3.75VVNKK300 pKa = 9.45FYY302 pKa = 11.39
Molecular weight: 35.57 kDa
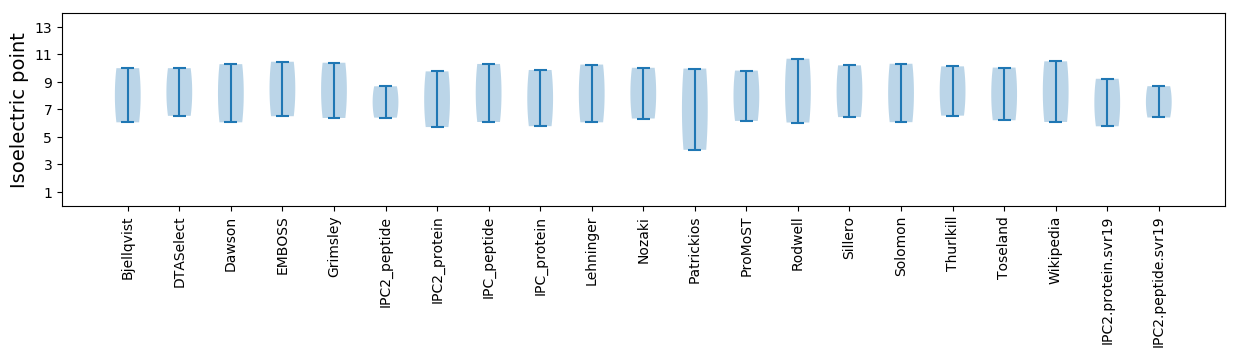
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J462|A0A075J462_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-4 OX=1519393 PE=3 SV=1
MM1 pKa = 7.69PKK3 pKa = 8.85YY4 pKa = 10.5AKK6 pKa = 9.73RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 7.23YY10 pKa = 8.7TNRR13 pKa = 11.84AAKK16 pKa = 9.94AFKK19 pKa = 10.43KK20 pKa = 10.23SVPKK24 pKa = 10.46TSTKK28 pKa = 8.57TIRR31 pKa = 11.84RR32 pKa = 11.84IAVKK36 pKa = 10.14AIKK39 pKa = 10.2SQAEE43 pKa = 4.11TKK45 pKa = 10.18NYY47 pKa = 8.55YY48 pKa = 8.03YY49 pKa = 10.64HH50 pKa = 7.45WLVQNQSTAFPYY62 pKa = 10.51AHH64 pKa = 6.54NAFYY68 pKa = 10.44EE69 pKa = 4.28INQGVGDD76 pKa = 3.89NQRR79 pKa = 11.84IGDD82 pKa = 3.88KK83 pKa = 10.74VYY85 pKa = 11.02VSRR88 pKa = 11.84LTYY91 pKa = 10.84KK92 pKa = 10.73LGFTIDD98 pKa = 3.75SNFFAPFGVRR108 pKa = 11.84GDD110 pKa = 3.33IYY112 pKa = 11.05VRR114 pKa = 11.84CLLVQNKK121 pKa = 8.63ARR123 pKa = 11.84QNNGITGFLPTWGGSVLMKK142 pKa = 10.35SGYY145 pKa = 8.16FQNGIVDD152 pKa = 3.99KK153 pKa = 10.94EE154 pKa = 4.21QFTVLRR160 pKa = 11.84DD161 pKa = 3.81RR162 pKa = 11.84NFHH165 pKa = 6.74LSPGNAINGKK175 pKa = 4.98QTYY178 pKa = 8.66YY179 pKa = 11.02RR180 pKa = 11.84NISVRR185 pKa = 11.84WAKK188 pKa = 10.49NIQYY192 pKa = 10.87DD193 pKa = 3.9SDD195 pKa = 3.43NGGYY199 pKa = 8.97MKK201 pKa = 10.74FKK203 pKa = 9.47NTYY206 pKa = 9.8LIYY209 pKa = 10.31QIYY212 pKa = 10.18SPDD215 pKa = 3.58GYY217 pKa = 7.85TTGGFTTNIVHH228 pKa = 7.25NIQFKK233 pKa = 10.89DD234 pKa = 3.43LL235 pKa = 3.76
MM1 pKa = 7.69PKK3 pKa = 8.85YY4 pKa = 10.5AKK6 pKa = 9.73RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 7.23YY10 pKa = 8.7TNRR13 pKa = 11.84AAKK16 pKa = 9.94AFKK19 pKa = 10.43KK20 pKa = 10.23SVPKK24 pKa = 10.46TSTKK28 pKa = 8.57TIRR31 pKa = 11.84RR32 pKa = 11.84IAVKK36 pKa = 10.14AIKK39 pKa = 10.2SQAEE43 pKa = 4.11TKK45 pKa = 10.18NYY47 pKa = 8.55YY48 pKa = 8.03YY49 pKa = 10.64HH50 pKa = 7.45WLVQNQSTAFPYY62 pKa = 10.51AHH64 pKa = 6.54NAFYY68 pKa = 10.44EE69 pKa = 4.28INQGVGDD76 pKa = 3.89NQRR79 pKa = 11.84IGDD82 pKa = 3.88KK83 pKa = 10.74VYY85 pKa = 11.02VSRR88 pKa = 11.84LTYY91 pKa = 10.84KK92 pKa = 10.73LGFTIDD98 pKa = 3.75SNFFAPFGVRR108 pKa = 11.84GDD110 pKa = 3.33IYY112 pKa = 11.05VRR114 pKa = 11.84CLLVQNKK121 pKa = 8.63ARR123 pKa = 11.84QNNGITGFLPTWGGSVLMKK142 pKa = 10.35SGYY145 pKa = 8.16FQNGIVDD152 pKa = 3.99KK153 pKa = 10.94EE154 pKa = 4.21QFTVLRR160 pKa = 11.84DD161 pKa = 3.81RR162 pKa = 11.84NFHH165 pKa = 6.74LSPGNAINGKK175 pKa = 4.98QTYY178 pKa = 8.66YY179 pKa = 11.02RR180 pKa = 11.84NISVRR185 pKa = 11.84WAKK188 pKa = 10.49NIQYY192 pKa = 10.87DD193 pKa = 3.9SDD195 pKa = 3.43NGGYY199 pKa = 8.97MKK201 pKa = 10.74FKK203 pKa = 9.47NTYY206 pKa = 9.8LIYY209 pKa = 10.31QIYY212 pKa = 10.18SPDD215 pKa = 3.58GYY217 pKa = 7.85TTGGFTTNIVHH228 pKa = 7.25NIQFKK233 pKa = 10.89DD234 pKa = 3.43LL235 pKa = 3.76
Molecular weight: 27.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
537 |
235 |
302 |
268.5 |
31.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.773 ± 0.116 | 1.304 ± 0.55 |
6.518 ± 1.418 | 4.097 ± 1.768 |
6.704 ± 0.468 | 5.587 ± 1.566 |
2.98 ± 0.801 | 6.704 ± 0.066 |
9.125 ± 0.118 | 5.4 ± 0.451 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.49 ± 0.134 | 7.263 ± 0.783 |
3.724 ± 0.468 | 3.911 ± 1.017 |
4.469 ± 0.933 | 4.469 ± 0.399 |
6.518 ± 0.449 | 5.773 ± 0.116 |
2.048 ± 0.484 | 6.145 ± 1.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
