
Metschnikowia bicuspidata var. bicuspidata NRRL YB-4993
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae; Metschnikowia; Metschnikowia bicuspidata; Metschnikowia bicuspidata var. bicuspidata
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
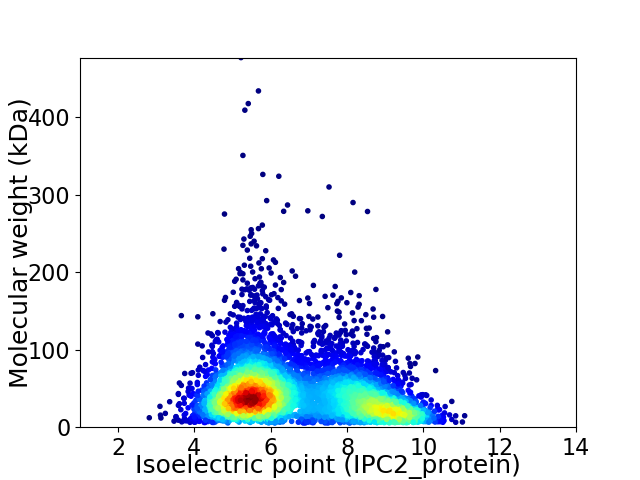
Virtual 2D-PAGE plot for 5827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A0H6J0|A0A1A0H6J0_9ASCO Putative transferase CAF17 mitochondrial OS=Metschnikowia bicuspidata var. bicuspidata NRRL YB-4993 OX=869754 GN=METBIDRAFT_33194 PE=3 SV=1
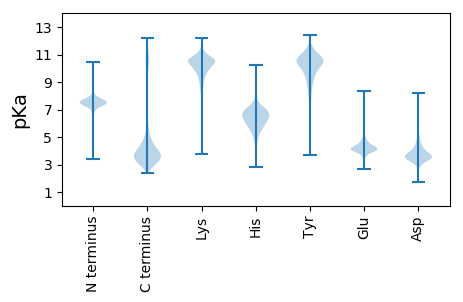
MM1 pKa = 7.65RR2 pKa = 11.84LSVLAWVATVLAGYY16 pKa = 10.67APFEE20 pKa = 4.3IDD22 pKa = 3.72CPEE25 pKa = 4.09GNLLRR30 pKa = 11.84LANSLSEE37 pKa = 4.54EE38 pKa = 4.06EE39 pKa = 4.36NDD41 pKa = 3.63WVTEE45 pKa = 3.98RR46 pKa = 11.84HH47 pKa = 5.78KK48 pKa = 11.03VTDD51 pKa = 3.61SYY53 pKa = 11.97LEE55 pKa = 4.13TFLNDD60 pKa = 3.74AMLDD64 pKa = 3.79DD65 pKa = 5.31FDD67 pKa = 6.06ASSFLRR73 pKa = 11.84NTTNNRR79 pKa = 11.84SITIGLAFSGGGYY92 pKa = 9.95RR93 pKa = 11.84AMLAGAGQLSALDD106 pKa = 3.65NRR108 pKa = 11.84TEE110 pKa = 4.07GAHH113 pKa = 6.87AEE115 pKa = 4.18GLGGLLQASTYY126 pKa = 10.9LVGLSGGSWLVGSVANNNFTSIQDD150 pKa = 3.35IVDD153 pKa = 3.57NKK155 pKa = 10.19IDD157 pKa = 3.51IWDD160 pKa = 3.82LEE162 pKa = 4.29YY163 pKa = 11.22SVVNMGGWNPIEE175 pKa = 4.24AYY177 pKa = 9.87QYY179 pKa = 11.61YY180 pKa = 10.29KK181 pKa = 10.95DD182 pKa = 4.93LYY184 pKa = 10.55DD185 pKa = 4.43DD186 pKa = 3.98VSAKK190 pKa = 10.21EE191 pKa = 3.82DD192 pKa = 3.3AGFDD196 pKa = 3.83VSLTDD201 pKa = 2.89TWGRR205 pKa = 11.84ALSHH209 pKa = 6.17QFFTEE214 pKa = 3.8QSDD217 pKa = 3.79YY218 pKa = 11.63GEE220 pKa = 4.62NILWSGIQDD229 pKa = 3.07IPEE232 pKa = 4.37FADD235 pKa = 3.19HH236 pKa = 6.89EE237 pKa = 4.76MPFPIVVADD246 pKa = 3.59GRR248 pKa = 11.84TPGTFVISGNSTVFEE263 pKa = 4.22FNPFEE268 pKa = 4.27MGSWDD273 pKa = 3.38PSLYY277 pKa = 10.95AFTKK281 pKa = 9.48TKK283 pKa = 10.92YY284 pKa = 10.39LGTDD288 pKa = 2.96VSDD291 pKa = 3.85GVPVNGSCIGGFDD304 pKa = 3.14NSGFVMGTSSSLFNQFVLQINTTSLSSTIQSLISLFLEE342 pKa = 5.92DD343 pKa = 5.02ISQDD347 pKa = 3.53EE348 pKa = 4.29NDD350 pKa = 3.33IALYY354 pKa = 10.15KK355 pKa = 10.36PNPFQNFSEE364 pKa = 4.67AGVDD368 pKa = 4.44SIVQNDD374 pKa = 3.74TLYY377 pKa = 11.27LVDD380 pKa = 5.32GGEE383 pKa = 4.33DD384 pKa = 3.43LQNIPLYY391 pKa = 10.11PLLQHH396 pKa = 6.22PRR398 pKa = 11.84EE399 pKa = 3.93VDD401 pKa = 3.57VIFAYY406 pKa = 10.55DD407 pKa = 3.46NSADD411 pKa = 3.94TDD413 pKa = 4.15EE414 pKa = 4.55NWPDD418 pKa = 3.51GASMVASYY426 pKa = 10.61QRR428 pKa = 11.84QFLEE432 pKa = 4.16QGNGTIFPYY441 pKa = 10.79VPDD444 pKa = 3.59VNSFRR449 pKa = 11.84NLNLTAKK456 pKa = 10.03PAFFGCNARR465 pKa = 11.84NLSSLLADD473 pKa = 5.1DD474 pKa = 5.4ADD476 pKa = 4.06EE477 pKa = 5.36DD478 pKa = 4.45DD479 pKa = 5.64VYY481 pKa = 11.5DD482 pKa = 4.1SPLIVYY488 pKa = 7.71TANRR492 pKa = 11.84PFSFHH497 pKa = 7.16SNTSTFQLSYY507 pKa = 10.9EE508 pKa = 4.25DD509 pKa = 4.27PEE511 pKa = 4.25KK512 pKa = 11.14LSMIRR517 pKa = 11.84NGFEE521 pKa = 3.61TASRR525 pKa = 11.84LNMTLDD531 pKa = 4.71DD532 pKa = 3.67EE533 pKa = 4.79WKK535 pKa = 8.63TCVACAIIRR544 pKa = 11.84RR545 pKa = 11.84EE546 pKa = 3.94QEE548 pKa = 3.65RR549 pKa = 11.84QGIEE553 pKa = 3.89QSDD556 pKa = 3.61QCKK559 pKa = 9.64QCFEE563 pKa = 4.45EE564 pKa = 4.6YY565 pKa = 10.17CWDD568 pKa = 3.58GTIDD572 pKa = 4.1DD573 pKa = 4.75SAPGVNFTTEE583 pKa = 3.73GTTDD587 pKa = 3.49DD588 pKa = 4.9AEE590 pKa = 4.37NTGNVSTSFGYY601 pKa = 10.5SLIRR605 pKa = 11.84SNKK608 pKa = 9.68NYY610 pKa = 10.93ALLQALGYY618 pKa = 10.12GLLAVVVCTYY628 pKa = 10.99SALL631 pKa = 3.72
MM1 pKa = 7.65RR2 pKa = 11.84LSVLAWVATVLAGYY16 pKa = 10.67APFEE20 pKa = 4.3IDD22 pKa = 3.72CPEE25 pKa = 4.09GNLLRR30 pKa = 11.84LANSLSEE37 pKa = 4.54EE38 pKa = 4.06EE39 pKa = 4.36NDD41 pKa = 3.63WVTEE45 pKa = 3.98RR46 pKa = 11.84HH47 pKa = 5.78KK48 pKa = 11.03VTDD51 pKa = 3.61SYY53 pKa = 11.97LEE55 pKa = 4.13TFLNDD60 pKa = 3.74AMLDD64 pKa = 3.79DD65 pKa = 5.31FDD67 pKa = 6.06ASSFLRR73 pKa = 11.84NTTNNRR79 pKa = 11.84SITIGLAFSGGGYY92 pKa = 9.95RR93 pKa = 11.84AMLAGAGQLSALDD106 pKa = 3.65NRR108 pKa = 11.84TEE110 pKa = 4.07GAHH113 pKa = 6.87AEE115 pKa = 4.18GLGGLLQASTYY126 pKa = 10.9LVGLSGGSWLVGSVANNNFTSIQDD150 pKa = 3.35IVDD153 pKa = 3.57NKK155 pKa = 10.19IDD157 pKa = 3.51IWDD160 pKa = 3.82LEE162 pKa = 4.29YY163 pKa = 11.22SVVNMGGWNPIEE175 pKa = 4.24AYY177 pKa = 9.87QYY179 pKa = 11.61YY180 pKa = 10.29KK181 pKa = 10.95DD182 pKa = 4.93LYY184 pKa = 10.55DD185 pKa = 4.43DD186 pKa = 3.98VSAKK190 pKa = 10.21EE191 pKa = 3.82DD192 pKa = 3.3AGFDD196 pKa = 3.83VSLTDD201 pKa = 2.89TWGRR205 pKa = 11.84ALSHH209 pKa = 6.17QFFTEE214 pKa = 3.8QSDD217 pKa = 3.79YY218 pKa = 11.63GEE220 pKa = 4.62NILWSGIQDD229 pKa = 3.07IPEE232 pKa = 4.37FADD235 pKa = 3.19HH236 pKa = 6.89EE237 pKa = 4.76MPFPIVVADD246 pKa = 3.59GRR248 pKa = 11.84TPGTFVISGNSTVFEE263 pKa = 4.22FNPFEE268 pKa = 4.27MGSWDD273 pKa = 3.38PSLYY277 pKa = 10.95AFTKK281 pKa = 9.48TKK283 pKa = 10.92YY284 pKa = 10.39LGTDD288 pKa = 2.96VSDD291 pKa = 3.85GVPVNGSCIGGFDD304 pKa = 3.14NSGFVMGTSSSLFNQFVLQINTTSLSSTIQSLISLFLEE342 pKa = 5.92DD343 pKa = 5.02ISQDD347 pKa = 3.53EE348 pKa = 4.29NDD350 pKa = 3.33IALYY354 pKa = 10.15KK355 pKa = 10.36PNPFQNFSEE364 pKa = 4.67AGVDD368 pKa = 4.44SIVQNDD374 pKa = 3.74TLYY377 pKa = 11.27LVDD380 pKa = 5.32GGEE383 pKa = 4.33DD384 pKa = 3.43LQNIPLYY391 pKa = 10.11PLLQHH396 pKa = 6.22PRR398 pKa = 11.84EE399 pKa = 3.93VDD401 pKa = 3.57VIFAYY406 pKa = 10.55DD407 pKa = 3.46NSADD411 pKa = 3.94TDD413 pKa = 4.15EE414 pKa = 4.55NWPDD418 pKa = 3.51GASMVASYY426 pKa = 10.61QRR428 pKa = 11.84QFLEE432 pKa = 4.16QGNGTIFPYY441 pKa = 10.79VPDD444 pKa = 3.59VNSFRR449 pKa = 11.84NLNLTAKK456 pKa = 10.03PAFFGCNARR465 pKa = 11.84NLSSLLADD473 pKa = 5.1DD474 pKa = 5.4ADD476 pKa = 4.06EE477 pKa = 5.36DD478 pKa = 4.45DD479 pKa = 5.64VYY481 pKa = 11.5DD482 pKa = 4.1SPLIVYY488 pKa = 7.71TANRR492 pKa = 11.84PFSFHH497 pKa = 7.16SNTSTFQLSYY507 pKa = 10.9EE508 pKa = 4.25DD509 pKa = 4.27PEE511 pKa = 4.25KK512 pKa = 11.14LSMIRR517 pKa = 11.84NGFEE521 pKa = 3.61TASRR525 pKa = 11.84LNMTLDD531 pKa = 4.71DD532 pKa = 3.67EE533 pKa = 4.79WKK535 pKa = 8.63TCVACAIIRR544 pKa = 11.84RR545 pKa = 11.84EE546 pKa = 3.94QEE548 pKa = 3.65RR549 pKa = 11.84QGIEE553 pKa = 3.89QSDD556 pKa = 3.61QCKK559 pKa = 9.64QCFEE563 pKa = 4.45EE564 pKa = 4.6YY565 pKa = 10.17CWDD568 pKa = 3.58GTIDD572 pKa = 4.1DD573 pKa = 4.75SAPGVNFTTEE583 pKa = 3.73GTTDD587 pKa = 3.49DD588 pKa = 4.9AEE590 pKa = 4.37NTGNVSTSFGYY601 pKa = 10.5SLIRR605 pKa = 11.84SNKK608 pKa = 9.68NYY610 pKa = 10.93ALLQALGYY618 pKa = 10.12GLLAVVVCTYY628 pKa = 10.99SALL631 pKa = 3.72
Molecular weight: 69.62 kDa
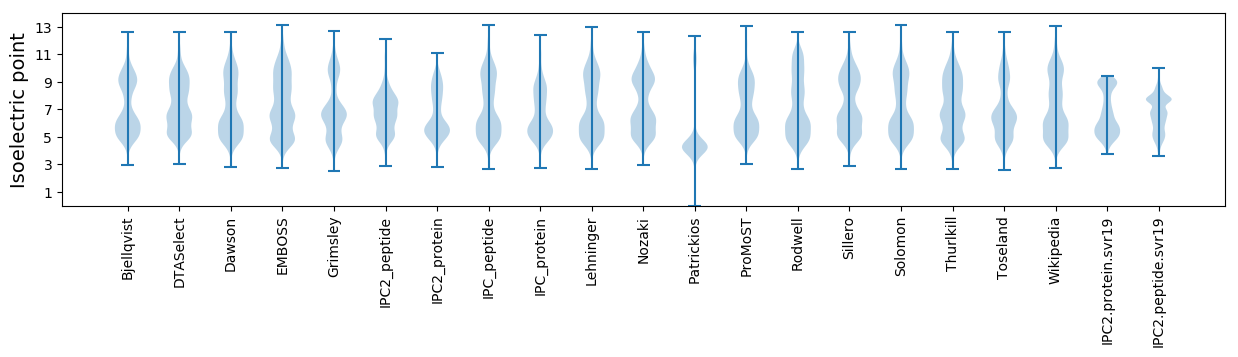
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A0HEQ5|A0A1A0HEQ5_9ASCO Uncharacterized protein OS=Metschnikowia bicuspidata var. bicuspidata NRRL YB-4993 OX=869754 GN=METBIDRAFT_82403 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.82PRR4 pKa = 11.84PVKK7 pKa = 10.6KK8 pKa = 10.16KK9 pKa = 10.04GRR11 pKa = 11.84SKK13 pKa = 10.72KK14 pKa = 9.5KK15 pKa = 9.2QGRR18 pKa = 11.84FNEE21 pKa = 4.1TRR23 pKa = 11.84PVQKK27 pKa = 10.02KK28 pKa = 9.87GRR30 pKa = 11.84FIKK33 pKa = 9.58TRR35 pKa = 11.84PLRR38 pKa = 11.84YY39 pKa = 9.92NKK41 pKa = 9.6VGQKK45 pKa = 10.05KK46 pKa = 7.85RR47 pKa = 11.84TAPIKK52 pKa = 10.24KK53 pKa = 9.1GRR55 pKa = 11.84SNITGPLQQNKK66 pKa = 9.29AASIKK71 pKa = 10.12QGRR74 pKa = 11.84TKK76 pKa = 10.67KK77 pKa = 9.09RR78 pKa = 11.84TAPTKK83 pKa = 9.87QGRR86 pKa = 11.84FNKK89 pKa = 9.5TGRR92 pKa = 11.84SKK94 pKa = 11.13KK95 pKa = 10.1KK96 pKa = 10.67GPLQKK101 pKa = 10.62KK102 pKa = 7.71KK103 pKa = 8.69TAPTKK108 pKa = 10.26AAPIKK113 pKa = 10.01QSRR116 pKa = 11.84FHH118 pKa = 6.4KK119 pKa = 9.49TRR121 pKa = 11.84AVLIKK126 pKa = 10.5PGLLL130 pKa = 3.47
MM1 pKa = 7.68KK2 pKa = 9.82PRR4 pKa = 11.84PVKK7 pKa = 10.6KK8 pKa = 10.16KK9 pKa = 10.04GRR11 pKa = 11.84SKK13 pKa = 10.72KK14 pKa = 9.5KK15 pKa = 9.2QGRR18 pKa = 11.84FNEE21 pKa = 4.1TRR23 pKa = 11.84PVQKK27 pKa = 10.02KK28 pKa = 9.87GRR30 pKa = 11.84FIKK33 pKa = 9.58TRR35 pKa = 11.84PLRR38 pKa = 11.84YY39 pKa = 9.92NKK41 pKa = 9.6VGQKK45 pKa = 10.05KK46 pKa = 7.85RR47 pKa = 11.84TAPIKK52 pKa = 10.24KK53 pKa = 9.1GRR55 pKa = 11.84SNITGPLQQNKK66 pKa = 9.29AASIKK71 pKa = 10.12QGRR74 pKa = 11.84TKK76 pKa = 10.67KK77 pKa = 9.09RR78 pKa = 11.84TAPTKK83 pKa = 9.87QGRR86 pKa = 11.84FNKK89 pKa = 9.5TGRR92 pKa = 11.84SKK94 pKa = 11.13KK95 pKa = 10.1KK96 pKa = 10.67GPLQKK101 pKa = 10.62KK102 pKa = 7.71KK103 pKa = 8.69TAPTKK108 pKa = 10.26AAPIKK113 pKa = 10.01QSRR116 pKa = 11.84FHH118 pKa = 6.4KK119 pKa = 9.49TRR121 pKa = 11.84AVLIKK126 pKa = 10.5PGLLL130 pKa = 3.47
Molecular weight: 14.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2545319 |
49 |
4188 |
436.8 |
48.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.797 ± 0.045 | 1.318 ± 0.012 |
5.739 ± 0.024 | 6.222 ± 0.03 |
4.482 ± 0.021 | 5.717 ± 0.033 |
2.454 ± 0.013 | 5.312 ± 0.029 |
6.089 ± 0.03 | 10.43 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.101 ± 0.011 | 4.405 ± 0.022 |
4.881 ± 0.028 | 3.844 ± 0.021 |
5.113 ± 0.024 | 8.185 ± 0.035 |
5.29 ± 0.018 | 6.413 ± 0.024 |
1.094 ± 0.009 | 3.113 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
