
Velvet tobacco mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
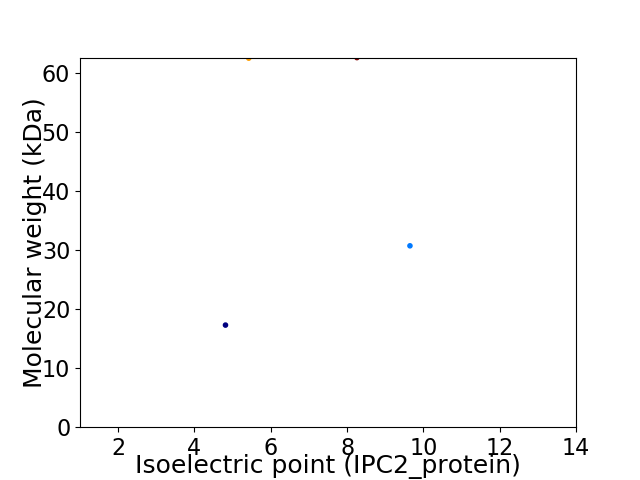
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
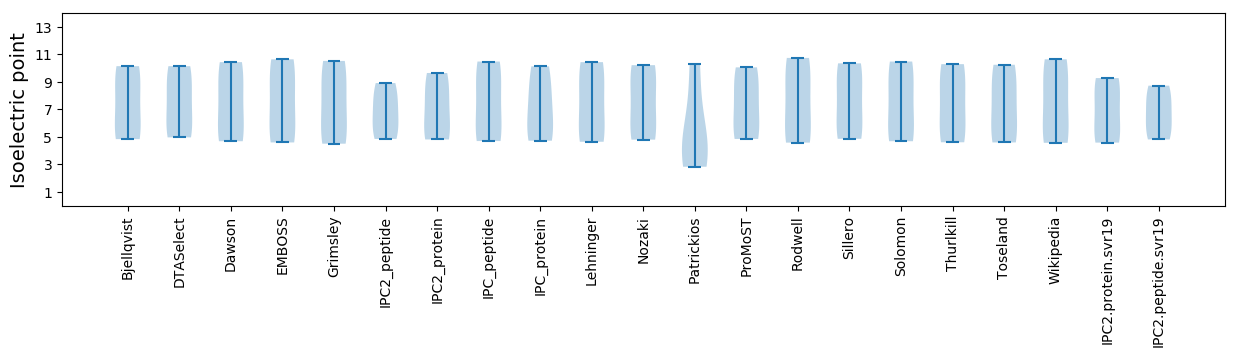
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2IZN1|E2IZN1_9VIRU N-terminal protein OS=Velvet tobacco mottle virus OX=12473 PE=4 SV=1
MM1 pKa = 7.71PSIDD5 pKa = 3.78VEE7 pKa = 4.43VEE9 pKa = 4.3KK10 pKa = 10.64ILHH13 pKa = 5.77LSNRR17 pKa = 11.84KK18 pKa = 8.79RR19 pKa = 11.84LRR21 pKa = 11.84SVCVARR27 pKa = 11.84KK28 pKa = 8.33KK29 pKa = 9.37TFVDD33 pKa = 4.34KK34 pKa = 10.72ISEE37 pKa = 4.32GFVCTLYY44 pKa = 11.2GEE46 pKa = 4.41YY47 pKa = 10.77DD48 pKa = 3.29HH49 pKa = 7.6DD50 pKa = 4.56YY51 pKa = 8.2VTSVHH56 pKa = 5.63LHH58 pKa = 5.87IVCSCGRR65 pKa = 11.84AFFDD69 pKa = 3.88FVEE72 pKa = 5.1FKK74 pKa = 10.59DD75 pKa = 3.18IKK77 pKa = 11.01LRR79 pKa = 11.84DD80 pKa = 3.68FQYY83 pKa = 10.89QSQCICPTRR92 pKa = 11.84NWSTVAYY99 pKa = 9.68TDD101 pKa = 3.91FTTVNCQVEE110 pKa = 4.48GCDD113 pKa = 3.33YY114 pKa = 11.44CEE116 pKa = 5.1GIEE119 pKa = 4.43SDD121 pKa = 3.74SDD123 pKa = 3.78SDD125 pKa = 3.89SEE127 pKa = 4.73AIIEE131 pKa = 4.1EE132 pKa = 4.18FLQKK136 pKa = 10.43FSEE139 pKa = 4.31VGISGSSSSPQTNN152 pKa = 2.6
MM1 pKa = 7.71PSIDD5 pKa = 3.78VEE7 pKa = 4.43VEE9 pKa = 4.3KK10 pKa = 10.64ILHH13 pKa = 5.77LSNRR17 pKa = 11.84KK18 pKa = 8.79RR19 pKa = 11.84LRR21 pKa = 11.84SVCVARR27 pKa = 11.84KK28 pKa = 8.33KK29 pKa = 9.37TFVDD33 pKa = 4.34KK34 pKa = 10.72ISEE37 pKa = 4.32GFVCTLYY44 pKa = 11.2GEE46 pKa = 4.41YY47 pKa = 10.77DD48 pKa = 3.29HH49 pKa = 7.6DD50 pKa = 4.56YY51 pKa = 8.2VTSVHH56 pKa = 5.63LHH58 pKa = 5.87IVCSCGRR65 pKa = 11.84AFFDD69 pKa = 3.88FVEE72 pKa = 5.1FKK74 pKa = 10.59DD75 pKa = 3.18IKK77 pKa = 11.01LRR79 pKa = 11.84DD80 pKa = 3.68FQYY83 pKa = 10.89QSQCICPTRR92 pKa = 11.84NWSTVAYY99 pKa = 9.68TDD101 pKa = 3.91FTTVNCQVEE110 pKa = 4.48GCDD113 pKa = 3.33YY114 pKa = 11.44CEE116 pKa = 5.1GIEE119 pKa = 4.43SDD121 pKa = 3.74SDD123 pKa = 3.78SDD125 pKa = 3.89SEE127 pKa = 4.73AIIEE131 pKa = 4.1EE132 pKa = 4.18FLQKK136 pKa = 10.43FSEE139 pKa = 4.31VGISGSSSSPQTNN152 pKa = 2.6
Molecular weight: 17.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2IZN3|E2IZN3_9VIRU Capsid protein OS=Velvet tobacco mottle virus OX=12473 GN=CP PE=3 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.89KK4 pKa = 8.99LTKK7 pKa = 10.61NQVKK11 pKa = 10.2QMIQATLPKK20 pKa = 9.12EE21 pKa = 3.99QTSARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84STQQGQSSTVMAPMAGAVIYY54 pKa = 9.99RR55 pKa = 11.84KK56 pKa = 9.99RR57 pKa = 11.84PMLINGRR64 pKa = 11.84SGVTVRR70 pKa = 11.84HH71 pKa = 5.6SEE73 pKa = 4.14VVLVVQSGTTNFSATSVTLAPNTFTWLAVQGSLYY107 pKa = 10.9SKK109 pKa = 9.65WRR111 pKa = 11.84WISLRR116 pKa = 11.84ATYY119 pKa = 10.55VPEE122 pKa = 4.34TASTTPGTVAMGFQYY137 pKa = 11.3DD138 pKa = 3.87NTDD141 pKa = 3.32VLPTGTAGMSSLHH154 pKa = 6.12GFVSGAPWSGFSGSKK169 pKa = 10.34LLAEE173 pKa = 4.93SPTTPIPAGAIATRR187 pKa = 11.84LDD189 pKa = 3.7CQNFGLKK196 pKa = 8.85WYY198 pKa = 9.3QYY200 pKa = 10.84KK201 pKa = 10.45SVIPAGDD208 pKa = 3.31SGNIYY213 pKa = 10.21IPAQLIVGTLGTGSTLRR230 pKa = 11.84YY231 pKa = 10.0GEE233 pKa = 3.94VHH235 pKa = 5.53IQYY238 pKa = 9.46EE239 pKa = 4.2IEE241 pKa = 4.77FIEE244 pKa = 4.24PLPPSVNTLRR254 pKa = 11.84DD255 pKa = 3.04IYY257 pKa = 11.42SRR259 pKa = 11.84MILSSASDD267 pKa = 3.34EE268 pKa = 4.36RR269 pKa = 11.84KK270 pKa = 10.29GEE272 pKa = 4.08KK273 pKa = 9.92SSQTLTDD280 pKa = 3.22RR281 pKa = 4.51
MM1 pKa = 7.53SKK3 pKa = 10.89KK4 pKa = 8.99LTKK7 pKa = 10.61NQVKK11 pKa = 10.2QMIQATLPKK20 pKa = 9.12EE21 pKa = 3.99QTSARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84STQQGQSSTVMAPMAGAVIYY54 pKa = 9.99RR55 pKa = 11.84KK56 pKa = 9.99RR57 pKa = 11.84PMLINGRR64 pKa = 11.84SGVTVRR70 pKa = 11.84HH71 pKa = 5.6SEE73 pKa = 4.14VVLVVQSGTTNFSATSVTLAPNTFTWLAVQGSLYY107 pKa = 10.9SKK109 pKa = 9.65WRR111 pKa = 11.84WISLRR116 pKa = 11.84ATYY119 pKa = 10.55VPEE122 pKa = 4.34TASTTPGTVAMGFQYY137 pKa = 11.3DD138 pKa = 3.87NTDD141 pKa = 3.32VLPTGTAGMSSLHH154 pKa = 6.12GFVSGAPWSGFSGSKK169 pKa = 10.34LLAEE173 pKa = 4.93SPTTPIPAGAIATRR187 pKa = 11.84LDD189 pKa = 3.7CQNFGLKK196 pKa = 8.85WYY198 pKa = 9.3QYY200 pKa = 10.84KK201 pKa = 10.45SVIPAGDD208 pKa = 3.31SGNIYY213 pKa = 10.21IPAQLIVGTLGTGSTLRR230 pKa = 11.84YY231 pKa = 10.0GEE233 pKa = 3.94VHH235 pKa = 5.53IQYY238 pKa = 9.46EE239 pKa = 4.2IEE241 pKa = 4.77FIEE244 pKa = 4.24PLPPSVNTLRR254 pKa = 11.84DD255 pKa = 3.04IYY257 pKa = 11.42SRR259 pKa = 11.84MILSSASDD267 pKa = 3.34EE268 pKa = 4.36RR269 pKa = 11.84KK270 pKa = 10.29GEE272 pKa = 4.08KK273 pKa = 9.92SSQTLTDD280 pKa = 3.22RR281 pKa = 4.51
Molecular weight: 30.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1553 |
152 |
572 |
388.3 |
43.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.504 ± 0.681 | 2.318 ± 0.88 |
4.701 ± 0.768 | 6.89 ± 0.875 |
3.67 ± 0.598 | 6.439 ± 0.449 |
1.674 ± 0.243 | 4.25 ± 0.574 |
5.666 ± 0.631 | 8.693 ± 0.836 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.189 ± 0.338 | 3.348 ± 0.287 |
5.473 ± 0.712 | 3.67 ± 0.387 |
6.182 ± 0.559 | 11.011 ± 0.602 |
5.409 ± 1.107 | 6.954 ± 1.14 |
1.996 ± 0.436 | 2.962 ± 0.372 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
