
Acidilobus sp. SCGC AC-742_E15
Taxonomy: cellular organisms; Archaea; TACK group; Crenarchaeota; Thermoprotei; Acidilobales; Acidilobaceae; Acidilobus; unclassified Acidilobus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
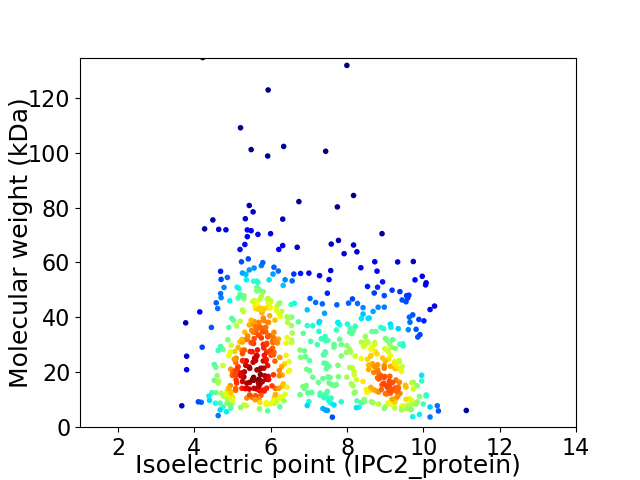
Virtual 2D-PAGE plot for 672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T9WZU2|A0A2T9WZU2_9CREN Site-specific DNA-methyltransferase (adenine-specific) (Fragment) OS=Acidilobus sp. SCGC AC-742_E15 OX=1987488 GN=DDW07_01715 PE=4 SV=1
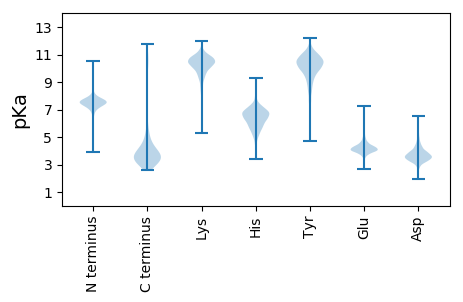
WW1 pKa = 7.23YY2 pKa = 8.92LAPHH6 pKa = 6.22VIKK9 pKa = 10.11WYY11 pKa = 10.1PNDD14 pKa = 3.42TGMAPISLAIALAQPSNVSGNAAITITITVASAYY48 pKa = 10.49VNQFTINGLMIAGAAIDD65 pKa = 4.19AVVDD69 pKa = 3.99DD70 pKa = 5.04VEE72 pKa = 5.13VGDD75 pKa = 4.42LFQAISQLFPQLGTTSTFTTEE96 pKa = 3.9GKK98 pKa = 9.56VAEE101 pKa = 4.52GNGVVVEE108 pKa = 5.19TITSTIIPGTADD120 pKa = 2.67IGQLYY125 pKa = 10.19IVGQVALVNWTAYY138 pKa = 8.27MLPLVSNTPVAWYY151 pKa = 10.43LSLQLTEE158 pKa = 5.53FKK160 pKa = 10.84AVEE163 pKa = 4.0NSGAVAPVIYY173 pKa = 9.69HH174 pKa = 5.43WWAYY178 pKa = 9.96DD179 pKa = 3.15PCINVSEE186 pKa = 4.33WAYY189 pKa = 8.85EE190 pKa = 4.03QQVLMQSPSYY200 pKa = 10.84FFGTGMNSTCPLPNTPAPVIWQDD223 pKa = 3.2YY224 pKa = 7.63ATNLTYY230 pKa = 9.2FTTKK234 pKa = 10.48NPNSQQQYY242 pKa = 8.16SYY244 pKa = 11.97VDD246 pKa = 3.65FFVNGGEE253 pKa = 4.13TLLGIGIDD261 pKa = 3.52AGSAIASIIGTVLMASQPEE280 pKa = 3.65IGAAVEE286 pKa = 4.61LIGALLDD293 pKa = 3.53SFQYY297 pKa = 10.52QSYY300 pKa = 10.18SVAVTVNNFLVSNTFPRR317 pKa = 11.84GYY319 pKa = 9.72YY320 pKa = 9.09VNVYY324 pKa = 9.54FSNLTPMYY332 pKa = 9.04TVSDD336 pKa = 3.45QGFTLPNEE344 pKa = 4.82LILINVTSS352 pKa = 3.61
WW1 pKa = 7.23YY2 pKa = 8.92LAPHH6 pKa = 6.22VIKK9 pKa = 10.11WYY11 pKa = 10.1PNDD14 pKa = 3.42TGMAPISLAIALAQPSNVSGNAAITITITVASAYY48 pKa = 10.49VNQFTINGLMIAGAAIDD65 pKa = 4.19AVVDD69 pKa = 3.99DD70 pKa = 5.04VEE72 pKa = 5.13VGDD75 pKa = 4.42LFQAISQLFPQLGTTSTFTTEE96 pKa = 3.9GKK98 pKa = 9.56VAEE101 pKa = 4.52GNGVVVEE108 pKa = 5.19TITSTIIPGTADD120 pKa = 2.67IGQLYY125 pKa = 10.19IVGQVALVNWTAYY138 pKa = 8.27MLPLVSNTPVAWYY151 pKa = 10.43LSLQLTEE158 pKa = 5.53FKK160 pKa = 10.84AVEE163 pKa = 4.0NSGAVAPVIYY173 pKa = 9.69HH174 pKa = 5.43WWAYY178 pKa = 9.96DD179 pKa = 3.15PCINVSEE186 pKa = 4.33WAYY189 pKa = 8.85EE190 pKa = 4.03QQVLMQSPSYY200 pKa = 10.84FFGTGMNSTCPLPNTPAPVIWQDD223 pKa = 3.2YY224 pKa = 7.63ATNLTYY230 pKa = 9.2FTTKK234 pKa = 10.48NPNSQQQYY242 pKa = 8.16SYY244 pKa = 11.97VDD246 pKa = 3.65FFVNGGEE253 pKa = 4.13TLLGIGIDD261 pKa = 3.52AGSAIASIIGTVLMASQPEE280 pKa = 3.65IGAAVEE286 pKa = 4.61LIGALLDD293 pKa = 3.53SFQYY297 pKa = 10.52QSYY300 pKa = 10.18SVAVTVNNFLVSNTFPRR317 pKa = 11.84GYY319 pKa = 9.72YY320 pKa = 9.09VNVYY324 pKa = 9.54FSNLTPMYY332 pKa = 9.04TVSDD336 pKa = 3.45QGFTLPNEE344 pKa = 4.82LILINVTSS352 pKa = 3.61
Molecular weight: 38.01 kDa
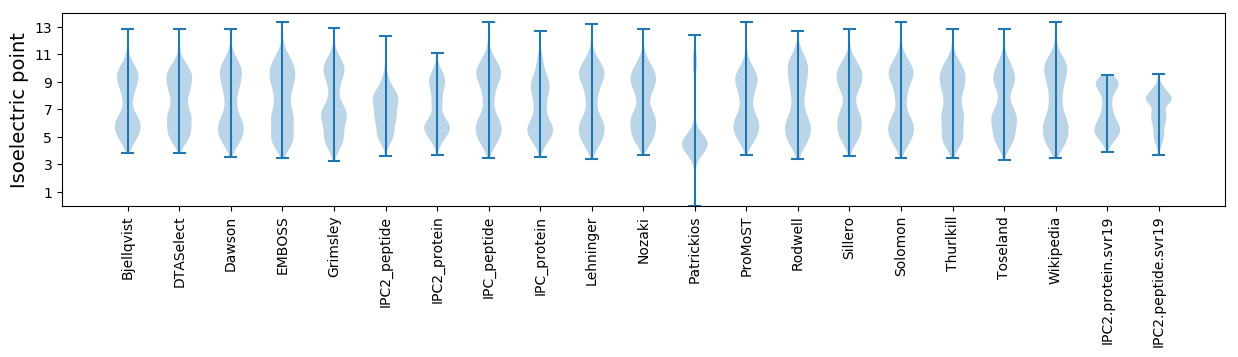
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T9X1A0|A0A2T9X1A0_9CREN Uncharacterized protein OS=Acidilobus sp. SCGC AC-742_E15 OX=1987488 GN=DDW07_01025 PE=4 SV=1
MM1 pKa = 7.42SRR3 pKa = 11.84HH4 pKa = 5.71KK5 pKa = 10.51PLARR9 pKa = 11.84KK10 pKa = 9.13LRR12 pKa = 11.84LARR15 pKa = 11.84ASKK18 pKa = 10.49SSQPVPAWVIVKK30 pKa = 7.32TLRR33 pKa = 11.84KK34 pKa = 8.6FTFNPKK40 pKa = 8.36RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.36WRR45 pKa = 11.84STKK48 pKa = 10.73LKK50 pKa = 9.86VV51 pKa = 3.04
MM1 pKa = 7.42SRR3 pKa = 11.84HH4 pKa = 5.71KK5 pKa = 10.51PLARR9 pKa = 11.84KK10 pKa = 9.13LRR12 pKa = 11.84LARR15 pKa = 11.84ASKK18 pKa = 10.49SSQPVPAWVIVKK30 pKa = 7.32TLRR33 pKa = 11.84KK34 pKa = 8.6FTFNPKK40 pKa = 8.36RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.36WRR45 pKa = 11.84STKK48 pKa = 10.73LKK50 pKa = 9.86VV51 pKa = 3.04
Molecular weight: 6.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
177325 |
32 |
1262 |
263.9 |
29.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.436 ± 0.105 | 0.658 ± 0.034 |
4.46 ± 0.079 | 6.937 ± 0.113 |
3.341 ± 0.057 | 8.034 ± 0.083 |
1.39 ± 0.035 | 5.282 ± 0.077 |
4.388 ± 0.11 | 11.154 ± 0.113 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.348 ± 0.043 | 2.619 ± 0.078 |
5.154 ± 0.083 | 2.15 ± 0.05 |
7.025 ± 0.142 | 6.627 ± 0.091 |
4.586 ± 0.101 | 9.287 ± 0.097 |
1.259 ± 0.045 | 3.864 ± 0.077 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
