
Odonata-associated circular virus-15
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.99
Get precalculated fractions of proteins
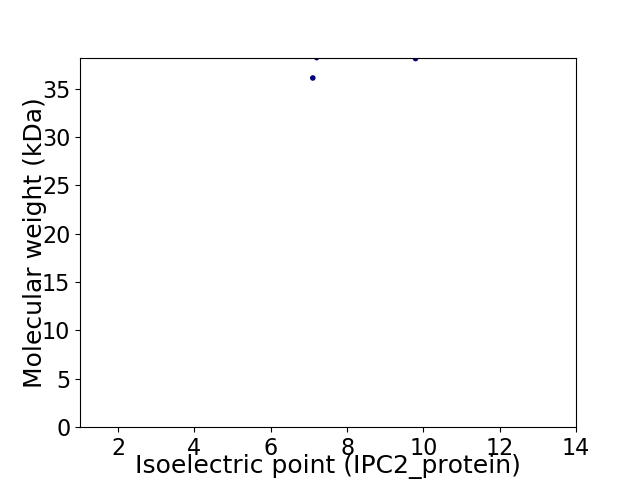
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGI9|A0A0B4UGI9_9VIRU Replication-associated protein OS=Odonata-associated circular virus-15 OX=1592115 PE=3 SV=1
MM1 pKa = 7.5SSNASSLSTSTNSFRR16 pKa = 11.84IQAKK20 pKa = 10.19SFFLTYY26 pKa = 8.6PQCSASKK33 pKa = 10.28QDD35 pKa = 4.34LKK37 pKa = 11.56NFLDD41 pKa = 3.65TKK43 pKa = 10.89GRR45 pKa = 11.84IVYY48 pKa = 8.78YY49 pKa = 10.67LIGQEE54 pKa = 3.78RR55 pKa = 11.84HH56 pKa = 5.46EE57 pKa = 5.39DD58 pKa = 3.95GNFHH62 pKa = 6.64LHH64 pKa = 6.92ALVTYY69 pKa = 8.17EE70 pKa = 4.34KK71 pKa = 10.17KK72 pKa = 10.68INVKK76 pKa = 9.02RR77 pKa = 11.84TTFFDD82 pKa = 4.09LNGFHH87 pKa = 7.47PNIQAAKK94 pKa = 10.06NLPALKK100 pKa = 10.42NYY102 pKa = 9.52ISKK105 pKa = 10.79EE106 pKa = 3.99DD107 pKa = 3.86IEE109 pKa = 4.76PLTSTVEE116 pKa = 4.04PEE118 pKa = 4.29EE119 pKa = 4.05EE120 pKa = 4.11DD121 pKa = 3.5NLYY124 pKa = 11.11DD125 pKa = 3.99LARR128 pKa = 11.84VTPEE132 pKa = 3.37EE133 pKa = 4.43NYY135 pKa = 10.46FEE137 pKa = 4.4ICRR140 pKa = 11.84KK141 pKa = 9.98RR142 pKa = 11.84KK143 pKa = 7.9VHH145 pKa = 6.14PSIISRR151 pKa = 11.84VPFMYY156 pKa = 10.63ANQAFLKK163 pKa = 9.44ILKK166 pKa = 9.34DD167 pKa = 3.44ASVNTIGEE175 pKa = 4.43EE176 pKa = 4.07YY177 pKa = 10.45QVIGTITSTVLQNLQLPTDD196 pKa = 3.87MTSLWVKK203 pKa = 10.19GPSGIGKK210 pKa = 5.86TTWALTVSNKK220 pKa = 9.2PALFVRR226 pKa = 11.84HH227 pKa = 6.8LDD229 pKa = 3.36TLRR232 pKa = 11.84EE233 pKa = 3.97FRR235 pKa = 11.84NGYY238 pKa = 9.51HH239 pKa = 5.6KK240 pKa = 9.59TIIFDD245 pKa = 3.81DD246 pKa = 3.89MSFQHH251 pKa = 6.94LPRR254 pKa = 11.84TAQIEE259 pKa = 4.38LVDD262 pKa = 4.61RR263 pKa = 11.84YY264 pKa = 10.24HH265 pKa = 6.83PQQIHH270 pKa = 4.91IRR272 pKa = 11.84YY273 pKa = 8.58AVVNIPPNIPKK284 pKa = 9.92IFLSNDD290 pKa = 3.41SIFTYY295 pKa = 10.67DD296 pKa = 3.03EE297 pKa = 4.85AIFRR301 pKa = 11.84RR302 pKa = 11.84LTLVNLEE309 pKa = 4.36FTDD312 pKa = 3.61QQ313 pKa = 3.28
MM1 pKa = 7.5SSNASSLSTSTNSFRR16 pKa = 11.84IQAKK20 pKa = 10.19SFFLTYY26 pKa = 8.6PQCSASKK33 pKa = 10.28QDD35 pKa = 4.34LKK37 pKa = 11.56NFLDD41 pKa = 3.65TKK43 pKa = 10.89GRR45 pKa = 11.84IVYY48 pKa = 8.78YY49 pKa = 10.67LIGQEE54 pKa = 3.78RR55 pKa = 11.84HH56 pKa = 5.46EE57 pKa = 5.39DD58 pKa = 3.95GNFHH62 pKa = 6.64LHH64 pKa = 6.92ALVTYY69 pKa = 8.17EE70 pKa = 4.34KK71 pKa = 10.17KK72 pKa = 10.68INVKK76 pKa = 9.02RR77 pKa = 11.84TTFFDD82 pKa = 4.09LNGFHH87 pKa = 7.47PNIQAAKK94 pKa = 10.06NLPALKK100 pKa = 10.42NYY102 pKa = 9.52ISKK105 pKa = 10.79EE106 pKa = 3.99DD107 pKa = 3.86IEE109 pKa = 4.76PLTSTVEE116 pKa = 4.04PEE118 pKa = 4.29EE119 pKa = 4.05EE120 pKa = 4.11DD121 pKa = 3.5NLYY124 pKa = 11.11DD125 pKa = 3.99LARR128 pKa = 11.84VTPEE132 pKa = 3.37EE133 pKa = 4.43NYY135 pKa = 10.46FEE137 pKa = 4.4ICRR140 pKa = 11.84KK141 pKa = 9.98RR142 pKa = 11.84KK143 pKa = 7.9VHH145 pKa = 6.14PSIISRR151 pKa = 11.84VPFMYY156 pKa = 10.63ANQAFLKK163 pKa = 9.44ILKK166 pKa = 9.34DD167 pKa = 3.44ASVNTIGEE175 pKa = 4.43EE176 pKa = 4.07YY177 pKa = 10.45QVIGTITSTVLQNLQLPTDD196 pKa = 3.87MTSLWVKK203 pKa = 10.19GPSGIGKK210 pKa = 5.86TTWALTVSNKK220 pKa = 9.2PALFVRR226 pKa = 11.84HH227 pKa = 6.8LDD229 pKa = 3.36TLRR232 pKa = 11.84EE233 pKa = 3.97FRR235 pKa = 11.84NGYY238 pKa = 9.51HH239 pKa = 5.6KK240 pKa = 9.59TIIFDD245 pKa = 3.81DD246 pKa = 3.89MSFQHH251 pKa = 6.94LPRR254 pKa = 11.84TAQIEE259 pKa = 4.38LVDD262 pKa = 4.61RR263 pKa = 11.84YY264 pKa = 10.24HH265 pKa = 6.83PQQIHH270 pKa = 4.91IRR272 pKa = 11.84YY273 pKa = 8.58AVVNIPPNIPKK284 pKa = 9.92IFLSNDD290 pKa = 3.41SIFTYY295 pKa = 10.67DD296 pKa = 3.03EE297 pKa = 4.85AIFRR301 pKa = 11.84RR302 pKa = 11.84LTLVNLEE309 pKa = 4.36FTDD312 pKa = 3.61QQ313 pKa = 3.28
Molecular weight: 36.09 kDa
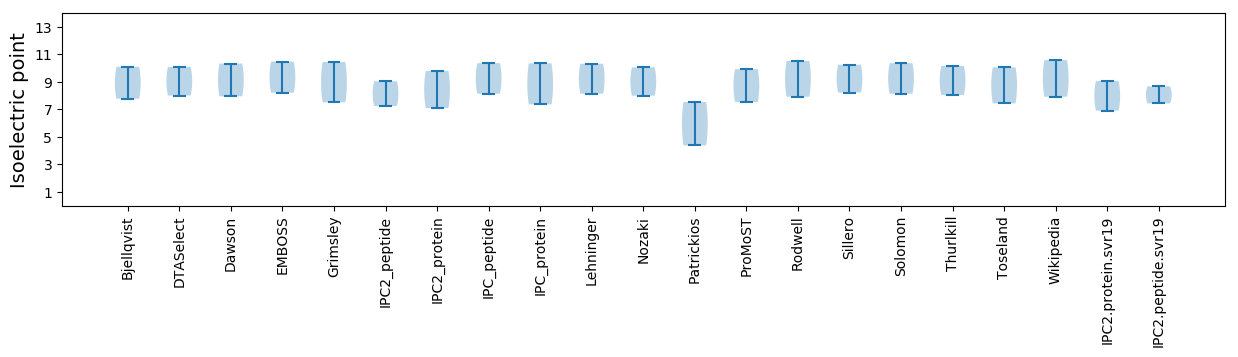
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGI9|A0A0B4UGI9_9VIRU Replication-associated protein OS=Odonata-associated circular virus-15 OX=1592115 PE=3 SV=1
MM1 pKa = 7.67IPLQMGYY8 pKa = 11.45SMVRR12 pKa = 11.84RR13 pKa = 11.84NARR16 pKa = 11.84AIGRR20 pKa = 11.84YY21 pKa = 9.25SGGAKK26 pKa = 9.0MAWRR30 pKa = 11.84LGQYY34 pKa = 10.05AGRR37 pKa = 11.84ALRR40 pKa = 11.84RR41 pKa = 11.84FRR43 pKa = 11.84RR44 pKa = 11.84GGGGGGSSSSAKK56 pKa = 10.2AKK58 pKa = 10.62AVGSLSEE65 pKa = 4.09QRR67 pKa = 11.84DD68 pKa = 3.44VTNLYY73 pKa = 9.61RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 9.2RR77 pKa = 11.84APRR80 pKa = 11.84RR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84AARR87 pKa = 11.84KK88 pKa = 5.34FTKK91 pKa = 10.23RR92 pKa = 11.84FMYY95 pKa = 10.25TLDD98 pKa = 3.2KK99 pKa = 8.79TQGMKK104 pKa = 9.3TCVITNAAQFSGSPTGLANGQQTIGITMYY133 pKa = 10.78GYY135 pKa = 8.0NTNSYY140 pKa = 10.68ASNIDD145 pKa = 3.68LGNGDD150 pKa = 4.36MPWIFARR157 pKa = 11.84EE158 pKa = 3.69NGAYY162 pKa = 6.85PTAASGTRR170 pKa = 11.84YY171 pKa = 9.95LRR173 pKa = 11.84FRR175 pKa = 11.84SCTMNYY181 pKa = 10.09SIQNTWEE188 pKa = 3.66QGVYY192 pKa = 8.93MDD194 pKa = 4.55IYY196 pKa = 10.73FVIARR201 pKa = 11.84KK202 pKa = 10.25SNGSTSDD209 pKa = 4.1PAAEE213 pKa = 4.05WNEE216 pKa = 3.79QIAIQNAGNMPTAITTNQYY235 pKa = 11.03YY236 pKa = 11.11GLTPFDD242 pKa = 3.55APGFGRR248 pKa = 11.84YY249 pKa = 7.41WLIKK253 pKa = 8.15SRR255 pKa = 11.84KK256 pKa = 8.75RR257 pKa = 11.84VFIQPAEE264 pKa = 4.07IYY266 pKa = 10.68SFQQRR271 pKa = 11.84DD272 pKa = 3.13AGNYY276 pKa = 7.46VLNMSDD282 pKa = 3.82VLDD285 pKa = 3.92IKK287 pKa = 10.88LKK289 pKa = 10.21QNLTEE294 pKa = 4.08GVILVFHH301 pKa = 6.65NPVTDD306 pKa = 3.42TVTTPGTIVPGGWQVQVTCTKK327 pKa = 8.98TYY329 pKa = 10.17HH330 pKa = 5.99YY331 pKa = 10.51SEE333 pKa = 4.99TSASTDD339 pKa = 3.5AIGVV343 pKa = 3.73
MM1 pKa = 7.67IPLQMGYY8 pKa = 11.45SMVRR12 pKa = 11.84RR13 pKa = 11.84NARR16 pKa = 11.84AIGRR20 pKa = 11.84YY21 pKa = 9.25SGGAKK26 pKa = 9.0MAWRR30 pKa = 11.84LGQYY34 pKa = 10.05AGRR37 pKa = 11.84ALRR40 pKa = 11.84RR41 pKa = 11.84FRR43 pKa = 11.84RR44 pKa = 11.84GGGGGGSSSSAKK56 pKa = 10.2AKK58 pKa = 10.62AVGSLSEE65 pKa = 4.09QRR67 pKa = 11.84DD68 pKa = 3.44VTNLYY73 pKa = 9.61RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 9.2RR77 pKa = 11.84APRR80 pKa = 11.84RR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84AARR87 pKa = 11.84KK88 pKa = 5.34FTKK91 pKa = 10.23RR92 pKa = 11.84FMYY95 pKa = 10.25TLDD98 pKa = 3.2KK99 pKa = 8.79TQGMKK104 pKa = 9.3TCVITNAAQFSGSPTGLANGQQTIGITMYY133 pKa = 10.78GYY135 pKa = 8.0NTNSYY140 pKa = 10.68ASNIDD145 pKa = 3.68LGNGDD150 pKa = 4.36MPWIFARR157 pKa = 11.84EE158 pKa = 3.69NGAYY162 pKa = 6.85PTAASGTRR170 pKa = 11.84YY171 pKa = 9.95LRR173 pKa = 11.84FRR175 pKa = 11.84SCTMNYY181 pKa = 10.09SIQNTWEE188 pKa = 3.66QGVYY192 pKa = 8.93MDD194 pKa = 4.55IYY196 pKa = 10.73FVIARR201 pKa = 11.84KK202 pKa = 10.25SNGSTSDD209 pKa = 4.1PAAEE213 pKa = 4.05WNEE216 pKa = 3.79QIAIQNAGNMPTAITTNQYY235 pKa = 11.03YY236 pKa = 11.11GLTPFDD242 pKa = 3.55APGFGRR248 pKa = 11.84YY249 pKa = 7.41WLIKK253 pKa = 8.15SRR255 pKa = 11.84KK256 pKa = 8.75RR257 pKa = 11.84VFIQPAEE264 pKa = 4.07IYY266 pKa = 10.68SFQQRR271 pKa = 11.84DD272 pKa = 3.13AGNYY276 pKa = 7.46VLNMSDD282 pKa = 3.82VLDD285 pKa = 3.92IKK287 pKa = 10.88LKK289 pKa = 10.21QNLTEE294 pKa = 4.08GVILVFHH301 pKa = 6.65NPVTDD306 pKa = 3.42TVTTPGTIVPGGWQVQVTCTKK327 pKa = 8.98TYY329 pKa = 10.17HH330 pKa = 5.99YY331 pKa = 10.51SEE333 pKa = 4.99TSASTDD339 pKa = 3.5AIGVV343 pKa = 3.73
Molecular weight: 38.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
313 |
343 |
328.0 |
37.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.165 ± 1.457 | 0.762 ± 0.087 |
4.268 ± 0.599 | 3.963 ± 1.269 |
4.726 ± 0.954 | 6.86 ± 2.601 |
1.829 ± 0.969 | 6.86 ± 0.8 |
5.03 ± 0.738 | 6.86 ± 1.707 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.824 | 6.098 ± 0.207 |
4.421 ± 0.49 | 4.878 ± 0.288 |
6.86 ± 1.241 | 7.012 ± 0.012 |
8.384 ± 0.282 | 5.335 ± 0.068 |
1.22 ± 0.412 | 5.03 ± 0.623 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
