
Capybara microvirus Cap1_SP_259
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
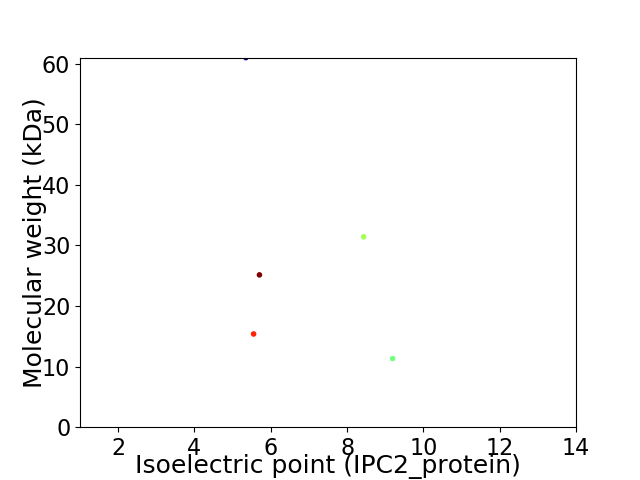
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4M0|A0A4P8W4M0_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_259 OX=2585419 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.48FSFKK6 pKa = 10.15RR7 pKa = 11.84SKK9 pKa = 9.24FTPHH13 pKa = 6.57SSRR16 pKa = 11.84IASLRR21 pKa = 11.84AGKK24 pKa = 9.92IYY26 pKa = 9.92PILVRR31 pKa = 11.84EE32 pKa = 4.55MYY34 pKa = 10.26PGDD37 pKa = 4.54TITIWHH43 pKa = 5.88QALLRR48 pKa = 11.84TAPMLFPLYY57 pKa = 10.91SPMKK61 pKa = 9.35LTFDD65 pKa = 3.61YY66 pKa = 10.04FYY68 pKa = 11.04SPNRR72 pKa = 11.84IVWKK76 pKa = 10.49GDD78 pKa = 3.2EE79 pKa = 4.36TKK81 pKa = 10.56DD82 pKa = 2.89WSAFITGSINGKK94 pKa = 10.04KK95 pKa = 9.65IEE97 pKa = 5.27DD98 pKa = 3.85PDD100 pKa = 4.09KK101 pKa = 11.1LPKK104 pKa = 10.21LPYY107 pKa = 8.66ITVPEE112 pKa = 4.55GGFGKK117 pKa = 10.58SSLADD122 pKa = 3.35YY123 pKa = 11.14LGFDD127 pKa = 3.57PSDD130 pKa = 3.45RR131 pKa = 11.84LEE133 pKa = 4.17GAKK136 pKa = 10.41LSALRR141 pKa = 11.84FRR143 pKa = 11.84HH144 pKa = 4.41YY145 pKa = 10.98QKK147 pKa = 10.59IISDD151 pKa = 4.01YY152 pKa = 10.71YY153 pKa = 11.07ADD155 pKa = 4.4EE156 pKa = 4.69NLSTMPEE163 pKa = 3.46IAYY166 pKa = 8.92TEE168 pKa = 4.67GEE170 pKa = 4.3DD171 pKa = 3.64TTTSLDD177 pKa = 3.57LFTRR181 pKa = 11.84CWNKK185 pKa = 10.54DD186 pKa = 3.06RR187 pKa = 11.84FTTASLDD194 pKa = 3.7PSSGEE199 pKa = 3.87SVMIPIGNSAPVVGNGNSLGLTNGVSDD226 pKa = 4.59FGLVSGQLYY235 pKa = 10.33INTDD239 pKa = 3.28RR240 pKa = 11.84EE241 pKa = 4.61GVLNTSQSTYY251 pKa = 11.2ALPASGLTEE260 pKa = 3.88RR261 pKa = 11.84AADD264 pKa = 3.57INNSTTALGVSKK276 pKa = 10.6DD277 pKa = 3.59SSKK280 pKa = 11.3SGLVADD286 pKa = 5.23LEE288 pKa = 4.44AATGIGVDD296 pKa = 3.58EE297 pKa = 4.46LHH299 pKa = 7.22FLVQLQRR306 pKa = 11.84LNQQDD311 pKa = 4.34AFWGTRR317 pKa = 11.84DD318 pKa = 3.6FEE320 pKa = 4.69CIYY323 pKa = 9.6THH325 pKa = 6.74FGVVIPDD332 pKa = 3.44TRR334 pKa = 11.84LQRR337 pKa = 11.84ATFLGRR343 pKa = 11.84SCVDD347 pKa = 3.17VNFSEE352 pKa = 4.81VLQTSQSTDD361 pKa = 2.6SSPQGNITGHH371 pKa = 6.69ALAVDD376 pKa = 4.07QNNPIHH382 pKa = 6.21FNCSEE387 pKa = 3.85HH388 pKa = 7.8GYY390 pKa = 10.3IFVLVNIQPIAQYY403 pKa = 10.31CQGIPRR409 pKa = 11.84DD410 pKa = 3.2AMYY413 pKa = 10.61RR414 pKa = 11.84DD415 pKa = 4.28RR416 pKa = 11.84YY417 pKa = 10.44SYY419 pKa = 8.69MWPEE423 pKa = 4.0FSEE426 pKa = 4.25TGQQEE431 pKa = 4.21IYY433 pKa = 10.41SAEE436 pKa = 4.13LYY438 pKa = 8.24ATRR441 pKa = 11.84QNIDD445 pKa = 2.76EE446 pKa = 4.36RR447 pKa = 11.84KK448 pKa = 8.91IFGYY452 pKa = 9.14EE453 pKa = 3.34PRR455 pKa = 11.84FNHH458 pKa = 6.46LRR460 pKa = 11.84FGDD463 pKa = 3.86KK464 pKa = 9.44TVHH467 pKa = 6.85GDD469 pKa = 3.57FRR471 pKa = 11.84DD472 pKa = 3.81NLSGYY477 pKa = 7.79HH478 pKa = 4.92TARR481 pKa = 11.84IFSEE485 pKa = 4.26EE486 pKa = 4.16PEE488 pKa = 4.18LNEE491 pKa = 3.62NFIRR495 pKa = 11.84ADD497 pKa = 3.06VTQRR501 pKa = 11.84MFAVNTQDD509 pKa = 4.01IDD511 pKa = 4.03QYY513 pKa = 8.24WLRR516 pKa = 11.84VEE518 pKa = 4.67FDD520 pKa = 3.14INHH523 pKa = 5.22VRR525 pKa = 11.84RR526 pKa = 11.84LPKK529 pKa = 9.82WPTPSAIGKK538 pKa = 8.66LFF540 pKa = 3.56
MM1 pKa = 7.42KK2 pKa = 10.48FSFKK6 pKa = 10.15RR7 pKa = 11.84SKK9 pKa = 9.24FTPHH13 pKa = 6.57SSRR16 pKa = 11.84IASLRR21 pKa = 11.84AGKK24 pKa = 9.92IYY26 pKa = 9.92PILVRR31 pKa = 11.84EE32 pKa = 4.55MYY34 pKa = 10.26PGDD37 pKa = 4.54TITIWHH43 pKa = 5.88QALLRR48 pKa = 11.84TAPMLFPLYY57 pKa = 10.91SPMKK61 pKa = 9.35LTFDD65 pKa = 3.61YY66 pKa = 10.04FYY68 pKa = 11.04SPNRR72 pKa = 11.84IVWKK76 pKa = 10.49GDD78 pKa = 3.2EE79 pKa = 4.36TKK81 pKa = 10.56DD82 pKa = 2.89WSAFITGSINGKK94 pKa = 10.04KK95 pKa = 9.65IEE97 pKa = 5.27DD98 pKa = 3.85PDD100 pKa = 4.09KK101 pKa = 11.1LPKK104 pKa = 10.21LPYY107 pKa = 8.66ITVPEE112 pKa = 4.55GGFGKK117 pKa = 10.58SSLADD122 pKa = 3.35YY123 pKa = 11.14LGFDD127 pKa = 3.57PSDD130 pKa = 3.45RR131 pKa = 11.84LEE133 pKa = 4.17GAKK136 pKa = 10.41LSALRR141 pKa = 11.84FRR143 pKa = 11.84HH144 pKa = 4.41YY145 pKa = 10.98QKK147 pKa = 10.59IISDD151 pKa = 4.01YY152 pKa = 10.71YY153 pKa = 11.07ADD155 pKa = 4.4EE156 pKa = 4.69NLSTMPEE163 pKa = 3.46IAYY166 pKa = 8.92TEE168 pKa = 4.67GEE170 pKa = 4.3DD171 pKa = 3.64TTTSLDD177 pKa = 3.57LFTRR181 pKa = 11.84CWNKK185 pKa = 10.54DD186 pKa = 3.06RR187 pKa = 11.84FTTASLDD194 pKa = 3.7PSSGEE199 pKa = 3.87SVMIPIGNSAPVVGNGNSLGLTNGVSDD226 pKa = 4.59FGLVSGQLYY235 pKa = 10.33INTDD239 pKa = 3.28RR240 pKa = 11.84EE241 pKa = 4.61GVLNTSQSTYY251 pKa = 11.2ALPASGLTEE260 pKa = 3.88RR261 pKa = 11.84AADD264 pKa = 3.57INNSTTALGVSKK276 pKa = 10.6DD277 pKa = 3.59SSKK280 pKa = 11.3SGLVADD286 pKa = 5.23LEE288 pKa = 4.44AATGIGVDD296 pKa = 3.58EE297 pKa = 4.46LHH299 pKa = 7.22FLVQLQRR306 pKa = 11.84LNQQDD311 pKa = 4.34AFWGTRR317 pKa = 11.84DD318 pKa = 3.6FEE320 pKa = 4.69CIYY323 pKa = 9.6THH325 pKa = 6.74FGVVIPDD332 pKa = 3.44TRR334 pKa = 11.84LQRR337 pKa = 11.84ATFLGRR343 pKa = 11.84SCVDD347 pKa = 3.17VNFSEE352 pKa = 4.81VLQTSQSTDD361 pKa = 2.6SSPQGNITGHH371 pKa = 6.69ALAVDD376 pKa = 4.07QNNPIHH382 pKa = 6.21FNCSEE387 pKa = 3.85HH388 pKa = 7.8GYY390 pKa = 10.3IFVLVNIQPIAQYY403 pKa = 10.31CQGIPRR409 pKa = 11.84DD410 pKa = 3.2AMYY413 pKa = 10.61RR414 pKa = 11.84DD415 pKa = 4.28RR416 pKa = 11.84YY417 pKa = 10.44SYY419 pKa = 8.69MWPEE423 pKa = 4.0FSEE426 pKa = 4.25TGQQEE431 pKa = 4.21IYY433 pKa = 10.41SAEE436 pKa = 4.13LYY438 pKa = 8.24ATRR441 pKa = 11.84QNIDD445 pKa = 2.76EE446 pKa = 4.36RR447 pKa = 11.84KK448 pKa = 8.91IFGYY452 pKa = 9.14EE453 pKa = 3.34PRR455 pKa = 11.84FNHH458 pKa = 6.46LRR460 pKa = 11.84FGDD463 pKa = 3.86KK464 pKa = 9.44TVHH467 pKa = 6.85GDD469 pKa = 3.57FRR471 pKa = 11.84DD472 pKa = 3.81NLSGYY477 pKa = 7.79HH478 pKa = 4.92TARR481 pKa = 11.84IFSEE485 pKa = 4.26EE486 pKa = 4.16PEE488 pKa = 4.18LNEE491 pKa = 3.62NFIRR495 pKa = 11.84ADD497 pKa = 3.06VTQRR501 pKa = 11.84MFAVNTQDD509 pKa = 4.01IDD511 pKa = 4.03QYY513 pKa = 8.24WLRR516 pKa = 11.84VEE518 pKa = 4.67FDD520 pKa = 3.14INHH523 pKa = 5.22VRR525 pKa = 11.84RR526 pKa = 11.84LPKK529 pKa = 9.82WPTPSAIGKK538 pKa = 8.66LFF540 pKa = 3.56
Molecular weight: 61.0 kDa
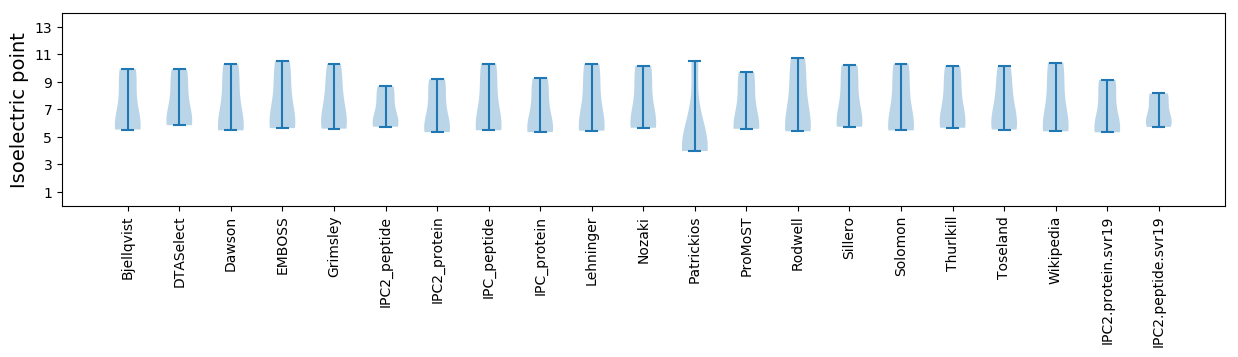
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W881|A0A4P8W881_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_259 OX=2585419 PE=4 SV=1
MM1 pKa = 7.54NISSFLSTRR10 pKa = 11.84KK11 pKa = 9.86AFRR14 pKa = 11.84EE15 pKa = 4.04FKK17 pKa = 10.83DD18 pKa = 3.47FGVFLDD24 pKa = 3.44VDD26 pKa = 3.61KK27 pKa = 11.45KK28 pKa = 10.48NRR30 pKa = 11.84CRR32 pKa = 11.84LMLRR36 pKa = 11.84HH37 pKa = 6.14IPSGRR42 pKa = 11.84VIEE45 pKa = 4.06LRR47 pKa = 11.84KK48 pKa = 9.47FVNVQSYY55 pKa = 7.83QRR57 pKa = 11.84FLVQGVFLRR66 pKa = 11.84LIADD70 pKa = 3.59SASVNKK76 pKa = 10.27ANHH79 pKa = 6.21SYY81 pKa = 11.25INVCNTLEE89 pKa = 3.96DD90 pKa = 4.74LIIKK94 pKa = 9.21EE95 pKa = 4.36SQQ97 pKa = 2.76
MM1 pKa = 7.54NISSFLSTRR10 pKa = 11.84KK11 pKa = 9.86AFRR14 pKa = 11.84EE15 pKa = 4.04FKK17 pKa = 10.83DD18 pKa = 3.47FGVFLDD24 pKa = 3.44VDD26 pKa = 3.61KK27 pKa = 11.45KK28 pKa = 10.48NRR30 pKa = 11.84CRR32 pKa = 11.84LMLRR36 pKa = 11.84HH37 pKa = 6.14IPSGRR42 pKa = 11.84VIEE45 pKa = 4.06LRR47 pKa = 11.84KK48 pKa = 9.47FVNVQSYY55 pKa = 7.83QRR57 pKa = 11.84FLVQGVFLRR66 pKa = 11.84LIADD70 pKa = 3.59SASVNKK76 pKa = 10.27ANHH79 pKa = 6.21SYY81 pKa = 11.25INVCNTLEE89 pKa = 3.96DD90 pKa = 4.74LIIKK94 pKa = 9.21EE95 pKa = 4.36SQQ97 pKa = 2.76
Molecular weight: 11.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1277 |
97 |
540 |
255.4 |
28.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.987 ± 2.329 | 1.41 ± 0.696 |
6.813 ± 0.53 | 4.777 ± 0.844 |
5.873 ± 0.815 | 6.265 ± 0.836 |
2.271 ± 0.304 | 5.012 ± 0.858 |
5.717 ± 0.997 | 8.771 ± 0.38 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.41 ± 0.223 | 5.951 ± 1.054 |
3.289 ± 1.052 | 3.994 ± 0.662 |
6.108 ± 0.95 | 8.457 ± 1.064 |
5.247 ± 1.007 | 5.09 ± 0.77 |
1.018 ± 0.313 | 4.542 ± 1.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
