
Ilumatobacter fluminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Acidimicrobiia; Acidimicrobiales; Ilumatobacteraceae; Ilumatobacter
Average proteome isoelectric point is 5.38
Get precalculated fractions of proteins
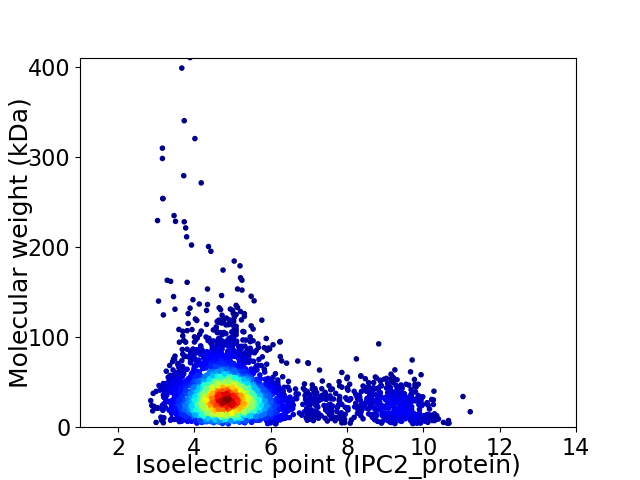
Virtual 2D-PAGE plot for 4258 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
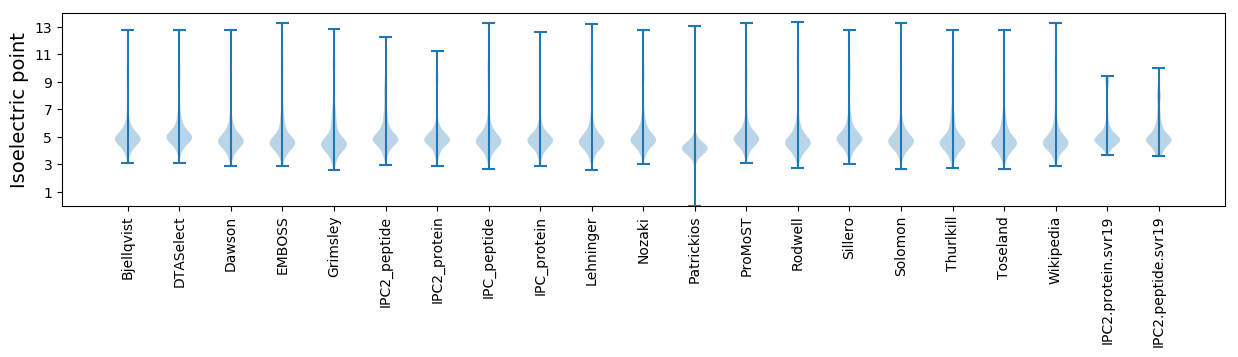
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7HXF4|A0A4R7HXF4_9ACTN NADH-quinone oxidoreductase subunit H OS=Ilumatobacter fluminis OX=467091 GN=nuoH PE=3 SV=1
MM1 pKa = 7.52TEE3 pKa = 4.44LANQMVMARR12 pKa = 11.84DD13 pKa = 3.76SDD15 pKa = 3.92QLDD18 pKa = 5.02LIDD21 pKa = 5.11DD22 pKa = 5.18CIDD25 pKa = 3.68TVPAIYY31 pKa = 10.51DD32 pKa = 3.52GTAPNCWTTCPPAEE46 pKa = 4.15RR47 pKa = 11.84TFHH50 pKa = 7.09LDD52 pKa = 2.9VARR55 pKa = 11.84NDD57 pKa = 3.45QLTNIQDD64 pKa = 3.96DD65 pKa = 4.25VWWRR69 pKa = 11.84TTLPVTYY76 pKa = 9.18VTDD79 pKa = 4.12DD80 pKa = 4.34GIIDD84 pKa = 3.61VTEE87 pKa = 3.88YY88 pKa = 10.37WSIRR92 pKa = 11.84QVDD95 pKa = 3.63DD96 pKa = 3.9RR97 pKa = 11.84FTVDD101 pKa = 2.95EE102 pKa = 5.29FEE104 pKa = 4.08IQTPIMQRR112 pKa = 11.84QSATDD117 pKa = 3.03ILTVYY122 pKa = 9.78FAHH125 pKa = 7.42IDD127 pKa = 3.68NEE129 pKa = 4.05NWAQAADD136 pKa = 4.53LLTQASSAPFGDD148 pKa = 4.76RR149 pKa = 11.84LDD151 pKa = 4.11LQQLGANSYY160 pKa = 10.32DD161 pKa = 3.37QGDD164 pKa = 3.72IADD167 pKa = 5.42ALARR171 pKa = 11.84WCADD175 pKa = 3.06GCDD178 pKa = 3.79TTPPSAAEE186 pKa = 3.79LTFTGGYY193 pKa = 9.74EE194 pKa = 4.03LTRR197 pKa = 11.84NAQTIRR203 pKa = 11.84VTWYY207 pKa = 7.94EE208 pKa = 3.96GAYY211 pKa = 9.97GIYY214 pKa = 10.25GPPLRR219 pKa = 5.81
MM1 pKa = 7.52TEE3 pKa = 4.44LANQMVMARR12 pKa = 11.84DD13 pKa = 3.76SDD15 pKa = 3.92QLDD18 pKa = 5.02LIDD21 pKa = 5.11DD22 pKa = 5.18CIDD25 pKa = 3.68TVPAIYY31 pKa = 10.51DD32 pKa = 3.52GTAPNCWTTCPPAEE46 pKa = 4.15RR47 pKa = 11.84TFHH50 pKa = 7.09LDD52 pKa = 2.9VARR55 pKa = 11.84NDD57 pKa = 3.45QLTNIQDD64 pKa = 3.96DD65 pKa = 4.25VWWRR69 pKa = 11.84TTLPVTYY76 pKa = 9.18VTDD79 pKa = 4.12DD80 pKa = 4.34GIIDD84 pKa = 3.61VTEE87 pKa = 3.88YY88 pKa = 10.37WSIRR92 pKa = 11.84QVDD95 pKa = 3.63DD96 pKa = 3.9RR97 pKa = 11.84FTVDD101 pKa = 2.95EE102 pKa = 5.29FEE104 pKa = 4.08IQTPIMQRR112 pKa = 11.84QSATDD117 pKa = 3.03ILTVYY122 pKa = 9.78FAHH125 pKa = 7.42IDD127 pKa = 3.68NEE129 pKa = 4.05NWAQAADD136 pKa = 4.53LLTQASSAPFGDD148 pKa = 4.76RR149 pKa = 11.84LDD151 pKa = 4.11LQQLGANSYY160 pKa = 10.32DD161 pKa = 3.37QGDD164 pKa = 3.72IADD167 pKa = 5.42ALARR171 pKa = 11.84WCADD175 pKa = 3.06GCDD178 pKa = 3.79TTPPSAAEE186 pKa = 3.79LTFTGGYY193 pKa = 9.74EE194 pKa = 4.03LTRR197 pKa = 11.84NAQTIRR203 pKa = 11.84VTWYY207 pKa = 7.94EE208 pKa = 3.96GAYY211 pKa = 9.97GIYY214 pKa = 10.25GPPLRR219 pKa = 5.81
Molecular weight: 24.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7I137|A0A4R7I137_9ACTN Lipoyl synthase OS=Ilumatobacter fluminis OX=467091 GN=lipA PE=3 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84PRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SDD9 pKa = 3.02PPTRR13 pKa = 11.84SRR15 pKa = 11.84GSRR18 pKa = 11.84TPPALTPTRR27 pKa = 11.84PHH29 pKa = 6.34RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 4.25HH41 pKa = 5.95RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.94DD47 pKa = 3.44PPLHH51 pKa = 6.35RR52 pKa = 11.84AHH54 pKa = 6.7GRR56 pKa = 11.84RR57 pKa = 11.84HH58 pKa = 3.6VRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84TGHH65 pKa = 5.81RR66 pKa = 11.84RR67 pKa = 11.84NHH69 pKa = 4.94RR70 pKa = 11.84HH71 pKa = 4.19RR72 pKa = 11.84HH73 pKa = 4.84HH74 pKa = 6.84RR75 pKa = 11.84HH76 pKa = 4.81HH77 pKa = 6.29HH78 pKa = 4.41HH79 pKa = 5.95HH80 pKa = 5.86RR81 pKa = 11.84HH82 pKa = 4.86RR83 pKa = 11.84RR84 pKa = 11.84SRR86 pKa = 11.84PRR88 pKa = 11.84QARR91 pKa = 11.84CHH93 pKa = 5.95RR94 pKa = 11.84PLRR97 pKa = 11.84RR98 pKa = 11.84HH99 pKa = 5.7RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84HH103 pKa = 4.79QRR105 pKa = 11.84DD106 pKa = 3.82SVHH109 pKa = 6.42LWPPTSPRR117 pKa = 11.84FGRR120 pKa = 11.84SICRR124 pKa = 11.84RR125 pKa = 11.84VACRR129 pKa = 11.84SLRR132 pKa = 11.84SRR134 pKa = 11.84RR135 pKa = 11.84SQNRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84VRR145 pKa = 11.84HH146 pKa = 5.79SSATSNDD153 pKa = 3.27CPPRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 4.85RR161 pKa = 11.84VRR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 4.05RR167 pKa = 11.84HH168 pKa = 4.42RR169 pKa = 11.84RR170 pKa = 11.84LDD172 pKa = 3.28RR173 pKa = 11.84GRR175 pKa = 11.84TVPPASRR182 pKa = 11.84RR183 pKa = 11.84QRR185 pKa = 11.84RR186 pKa = 11.84PVVPRR191 pKa = 11.84PTPGRR196 pKa = 11.84PGASSRR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84PLRR208 pKa = 11.84DD209 pKa = 3.1PARR212 pKa = 11.84RR213 pKa = 11.84PSGRR217 pKa = 11.84PASSDD222 pKa = 2.81GSAHH226 pKa = 6.56HH227 pKa = 6.61SPPGRR232 pKa = 11.84RR233 pKa = 11.84TWSIARR239 pKa = 11.84PHH241 pKa = 5.4PTPRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84SPPPPARR255 pKa = 11.84PRR257 pKa = 11.84HH258 pKa = 4.77RR259 pKa = 11.84HH260 pKa = 4.73FLSRR264 pKa = 11.84PPGLRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84DD272 pKa = 3.49RR273 pKa = 11.84ASSRR277 pKa = 3.37
MM1 pKa = 7.79RR2 pKa = 11.84PRR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84SDD9 pKa = 3.02PPTRR13 pKa = 11.84SRR15 pKa = 11.84GSRR18 pKa = 11.84TPPALTPTRR27 pKa = 11.84PHH29 pKa = 6.34RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 4.25HH41 pKa = 5.95RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.94DD47 pKa = 3.44PPLHH51 pKa = 6.35RR52 pKa = 11.84AHH54 pKa = 6.7GRR56 pKa = 11.84RR57 pKa = 11.84HH58 pKa = 3.6VRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84TGHH65 pKa = 5.81RR66 pKa = 11.84RR67 pKa = 11.84NHH69 pKa = 4.94RR70 pKa = 11.84HH71 pKa = 4.19RR72 pKa = 11.84HH73 pKa = 4.84HH74 pKa = 6.84RR75 pKa = 11.84HH76 pKa = 4.81HH77 pKa = 6.29HH78 pKa = 4.41HH79 pKa = 5.95HH80 pKa = 5.86RR81 pKa = 11.84HH82 pKa = 4.86RR83 pKa = 11.84RR84 pKa = 11.84SRR86 pKa = 11.84PRR88 pKa = 11.84QARR91 pKa = 11.84CHH93 pKa = 5.95RR94 pKa = 11.84PLRR97 pKa = 11.84RR98 pKa = 11.84HH99 pKa = 5.7RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84HH103 pKa = 4.79QRR105 pKa = 11.84DD106 pKa = 3.82SVHH109 pKa = 6.42LWPPTSPRR117 pKa = 11.84FGRR120 pKa = 11.84SICRR124 pKa = 11.84RR125 pKa = 11.84VACRR129 pKa = 11.84SLRR132 pKa = 11.84SRR134 pKa = 11.84RR135 pKa = 11.84SQNRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84VRR145 pKa = 11.84HH146 pKa = 5.79SSATSNDD153 pKa = 3.27CPPRR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 4.85RR161 pKa = 11.84VRR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84HH166 pKa = 4.05RR167 pKa = 11.84HH168 pKa = 4.42RR169 pKa = 11.84RR170 pKa = 11.84LDD172 pKa = 3.28RR173 pKa = 11.84GRR175 pKa = 11.84TVPPASRR182 pKa = 11.84RR183 pKa = 11.84QRR185 pKa = 11.84RR186 pKa = 11.84PVVPRR191 pKa = 11.84PTPGRR196 pKa = 11.84PGASSRR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 11.84PLRR208 pKa = 11.84DD209 pKa = 3.1PARR212 pKa = 11.84RR213 pKa = 11.84PSGRR217 pKa = 11.84PASSDD222 pKa = 2.81GSAHH226 pKa = 6.56HH227 pKa = 6.61SPPGRR232 pKa = 11.84RR233 pKa = 11.84TWSIARR239 pKa = 11.84PHH241 pKa = 5.4PTPRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84SPPPPARR255 pKa = 11.84PRR257 pKa = 11.84HH258 pKa = 4.77RR259 pKa = 11.84HH260 pKa = 4.73FLSRR264 pKa = 11.84PPGLRR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84DD272 pKa = 3.49RR273 pKa = 11.84ASSRR277 pKa = 3.37
Molecular weight: 33.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1475297 |
33 |
3915 |
346.5 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.18 ± 0.048 | 0.804 ± 0.011 |
7.852 ± 0.048 | 5.869 ± 0.037 |
3.142 ± 0.022 | 8.943 ± 0.036 |
2.141 ± 0.021 | 4.41 ± 0.028 |
1.605 ± 0.027 | 9.138 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.968 ± 0.022 | 2.095 ± 0.022 |
5.6 ± 0.029 | 2.546 ± 0.017 |
7.037 ± 0.054 | 5.467 ± 0.031 |
6.465 ± 0.055 | 9.248 ± 0.037 |
1.549 ± 0.018 | 1.94 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
