
Absidia repens
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Cunninghamellaceae; Absidia
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
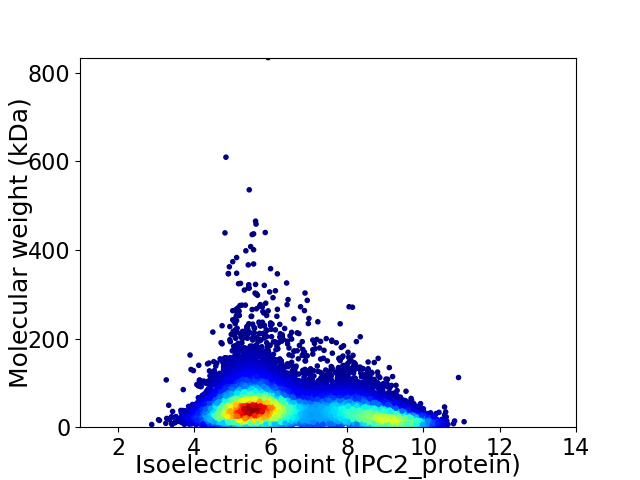
Virtual 2D-PAGE plot for 14767 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X2I7U2|A0A1X2I7U2_9FUNG CUE domain-containing protein OS=Absidia repens OX=90262 GN=BCR42DRAFT_421547 PE=4 SV=1
MM1 pKa = 7.38TSAFNSGIFKK11 pKa = 9.72TLVEE15 pKa = 4.31ANQRR19 pKa = 11.84SCDD22 pKa = 4.17LINSFVGSFDD32 pKa = 3.78FNRR35 pKa = 11.84NKK37 pKa = 10.77SNTLQEE43 pKa = 4.59LKK45 pKa = 10.51EE46 pKa = 4.18QTFHH50 pKa = 6.0TKK52 pKa = 10.45LYY54 pKa = 10.64INIQNTLPFIKK65 pKa = 10.44HH66 pKa = 6.53NIPKK70 pKa = 9.91AYY72 pKa = 10.34CEE74 pKa = 3.87LSYY77 pKa = 11.41EE78 pKa = 4.72NIVLWIQRR86 pKa = 11.84ALYY89 pKa = 9.12RR90 pKa = 11.84INVATFIRR98 pKa = 11.84FLEE101 pKa = 4.2LDD103 pKa = 3.72NIYY106 pKa = 10.99NAGKK110 pKa = 10.28FSLHH114 pKa = 6.29HH115 pKa = 5.72ICYY118 pKa = 10.19EE119 pKa = 3.91EE120 pKa = 4.01EE121 pKa = 4.04TRR123 pKa = 11.84SNDD126 pKa = 3.07VQAAFVDD133 pKa = 3.83LRR135 pKa = 11.84TRR137 pKa = 11.84LLATLVADD145 pKa = 5.79EE146 pKa = 5.38IDD148 pKa = 3.4TSSNEE153 pKa = 3.79NTSIEE158 pKa = 4.13MSPLMEE164 pKa = 4.13RR165 pKa = 11.84TLPLDD170 pKa = 3.84GSAFGLYY177 pKa = 9.86PEE179 pKa = 5.2EE180 pKa = 5.92DD181 pKa = 3.42DD182 pKa = 6.48DD183 pKa = 7.19FILEE187 pKa = 4.16LLKK190 pKa = 10.38QRR192 pKa = 11.84HH193 pKa = 5.1EE194 pKa = 4.79MITTLPDD201 pKa = 3.26SDD203 pKa = 5.16ALDD206 pKa = 4.08ALVTADD212 pKa = 4.04NLINVWKK219 pKa = 9.83VAIIASMYY227 pKa = 10.52RR228 pKa = 11.84MPALEE233 pKa = 5.17LPPGFPPNQSNDD245 pKa = 3.05DD246 pKa = 3.92VYY248 pKa = 11.13IKK250 pKa = 10.67QEE252 pKa = 3.96DD253 pKa = 4.33VLTDD257 pKa = 3.62VYY259 pKa = 11.53SDD261 pKa = 3.32AQEE264 pKa = 3.98YY265 pKa = 10.35QYY267 pKa = 11.52SQDD270 pKa = 3.04RR271 pKa = 11.84HH272 pKa = 5.75LSQVYY277 pKa = 9.21QLSQEE282 pKa = 4.31GEE284 pKa = 4.3SYY286 pKa = 10.99EE287 pKa = 4.78SNTHH291 pKa = 5.73GNTPIKK297 pKa = 10.68QEE299 pKa = 5.24DD300 pKa = 3.67DD301 pKa = 3.64DD302 pKa = 4.79NYY304 pKa = 11.5NGIAAQQYY312 pKa = 9.93GSTTFTTPCDD322 pKa = 3.6SGDD325 pKa = 4.26DD326 pKa = 4.01SDD328 pKa = 5.32HH329 pKa = 6.85SYY331 pKa = 11.13QGVEE335 pKa = 3.95YY336 pKa = 10.59LPNPVLATHH345 pKa = 7.41ILPGDD350 pKa = 3.68DD351 pKa = 3.72PEE353 pKa = 4.6FFEE356 pKa = 4.64GDD358 pKa = 3.54RR359 pKa = 11.84YY360 pKa = 11.13VLASPSLTEE369 pKa = 4.35HH370 pKa = 7.35DD371 pKa = 5.48LPGDD375 pKa = 4.01DD376 pKa = 6.02DD377 pKa = 5.52NDD379 pKa = 4.05FEE381 pKa = 6.29GDD383 pKa = 3.4QYY385 pKa = 11.86HH386 pKa = 7.21LARR389 pKa = 11.84PSFAEE394 pKa = 4.31NGLPDD399 pKa = 4.13NVEE402 pKa = 4.01KK403 pKa = 10.85DD404 pKa = 3.68DD405 pKa = 5.12EE406 pKa = 4.62NGQSHH411 pKa = 7.34LYY413 pKa = 9.76EE414 pKa = 4.66PPEE417 pKa = 4.23DD418 pKa = 3.72SNYY421 pKa = 10.57CEE423 pKa = 4.74DD424 pKa = 3.9QLGTLDD430 pKa = 4.38EE431 pKa = 4.49QNKK434 pKa = 9.86HH435 pKa = 6.47PDD437 pKa = 3.34TNINTDD443 pKa = 2.98TDD445 pKa = 3.51TDD447 pKa = 3.48TDD449 pKa = 3.9TNINITITINSDD461 pKa = 3.46SITNSNTNTMNVHH474 pKa = 5.57STDD477 pKa = 3.25KK478 pKa = 11.34AGDD481 pKa = 3.58GSEE484 pKa = 4.06GGGILEE490 pKa = 5.01EE491 pKa = 4.65NDD493 pKa = 3.86DD494 pKa = 4.0DD495 pKa = 5.9AGNQLEE501 pKa = 4.49EE502 pKa = 4.5SEE504 pKa = 4.93ADD506 pKa = 3.39HH507 pKa = 7.28DD508 pKa = 4.32HH509 pKa = 6.77GSVGTHH515 pKa = 5.44TPIIDD520 pKa = 3.95WNRR523 pKa = 11.84HH524 pKa = 4.78LAPSTAISDD533 pKa = 3.84PFSDD537 pKa = 4.67SEE539 pKa = 4.21IGHH542 pKa = 6.72DD543 pKa = 5.83LEE545 pKa = 7.27DD546 pKa = 4.16NDD548 pKa = 5.66DD549 pKa = 3.97NNEE552 pKa = 3.71FDD554 pKa = 4.98LFPAAHH560 pKa = 6.97EE561 pKa = 4.54DD562 pKa = 4.56DD563 pKa = 4.25YY564 pKa = 12.16AADD567 pKa = 3.7VNNMYY572 pKa = 7.76EE573 pKa = 4.4TTSPWNFDD581 pKa = 3.47DD582 pKa = 4.47ATEE585 pKa = 4.06FDD587 pKa = 3.96INQQYY592 pKa = 9.1TMVDD596 pKa = 4.24EE597 pKa = 4.51IAGDD601 pKa = 4.0DD602 pKa = 5.2DD603 pKa = 4.84IDD605 pKa = 4.28TPSEE609 pKa = 4.29MPTKK613 pKa = 10.73ADD615 pKa = 3.37DD616 pKa = 3.62TVTIDD621 pKa = 3.22THH623 pKa = 4.74TTAIDD628 pKa = 3.54NSSSSQAHH636 pKa = 5.85MDD638 pKa = 3.41STSVHH643 pKa = 6.6GISNQLEE650 pKa = 4.18SSPVDD655 pKa = 3.04THH657 pKa = 7.33LATEE661 pKa = 4.31AQNDD665 pKa = 3.9DD666 pKa = 4.08EE667 pKa = 5.12GADD670 pKa = 3.81DD671 pKa = 6.21LVDD674 pKa = 3.97EE675 pKa = 5.08YY676 pKa = 11.33EE677 pKa = 4.48DD678 pKa = 3.14VDD680 pKa = 3.87NRR682 pKa = 11.84VEE684 pKa = 3.97ASAEE688 pKa = 3.81ALEE691 pKa = 4.79NDD693 pKa = 4.5DD694 pKa = 4.99LTTTSPTLAGAHH706 pKa = 5.15VAGNDD711 pKa = 3.69ASDD714 pKa = 3.72SDD716 pKa = 4.01EE717 pKa = 4.65DD718 pKa = 5.97LIDD721 pKa = 3.94EE722 pKa = 5.41LEE724 pKa = 4.49NDD726 pKa = 4.46DD727 pKa = 5.98DD728 pKa = 6.63DD729 pKa = 6.4DD730 pKa = 6.01SEE732 pKa = 4.59YY733 pKa = 11.13QLDD736 pKa = 4.32PDD738 pKa = 4.35EE739 pKa = 5.27MDD741 pKa = 5.29DD742 pKa = 4.93SSSTNNSYY750 pKa = 10.71IEE752 pKa = 4.2TTGGEE757 pKa = 4.44GEE759 pKa = 4.29GRR761 pKa = 11.84NN762 pKa = 3.71
MM1 pKa = 7.38TSAFNSGIFKK11 pKa = 9.72TLVEE15 pKa = 4.31ANQRR19 pKa = 11.84SCDD22 pKa = 4.17LINSFVGSFDD32 pKa = 3.78FNRR35 pKa = 11.84NKK37 pKa = 10.77SNTLQEE43 pKa = 4.59LKK45 pKa = 10.51EE46 pKa = 4.18QTFHH50 pKa = 6.0TKK52 pKa = 10.45LYY54 pKa = 10.64INIQNTLPFIKK65 pKa = 10.44HH66 pKa = 6.53NIPKK70 pKa = 9.91AYY72 pKa = 10.34CEE74 pKa = 3.87LSYY77 pKa = 11.41EE78 pKa = 4.72NIVLWIQRR86 pKa = 11.84ALYY89 pKa = 9.12RR90 pKa = 11.84INVATFIRR98 pKa = 11.84FLEE101 pKa = 4.2LDD103 pKa = 3.72NIYY106 pKa = 10.99NAGKK110 pKa = 10.28FSLHH114 pKa = 6.29HH115 pKa = 5.72ICYY118 pKa = 10.19EE119 pKa = 3.91EE120 pKa = 4.01EE121 pKa = 4.04TRR123 pKa = 11.84SNDD126 pKa = 3.07VQAAFVDD133 pKa = 3.83LRR135 pKa = 11.84TRR137 pKa = 11.84LLATLVADD145 pKa = 5.79EE146 pKa = 5.38IDD148 pKa = 3.4TSSNEE153 pKa = 3.79NTSIEE158 pKa = 4.13MSPLMEE164 pKa = 4.13RR165 pKa = 11.84TLPLDD170 pKa = 3.84GSAFGLYY177 pKa = 9.86PEE179 pKa = 5.2EE180 pKa = 5.92DD181 pKa = 3.42DD182 pKa = 6.48DD183 pKa = 7.19FILEE187 pKa = 4.16LLKK190 pKa = 10.38QRR192 pKa = 11.84HH193 pKa = 5.1EE194 pKa = 4.79MITTLPDD201 pKa = 3.26SDD203 pKa = 5.16ALDD206 pKa = 4.08ALVTADD212 pKa = 4.04NLINVWKK219 pKa = 9.83VAIIASMYY227 pKa = 10.52RR228 pKa = 11.84MPALEE233 pKa = 5.17LPPGFPPNQSNDD245 pKa = 3.05DD246 pKa = 3.92VYY248 pKa = 11.13IKK250 pKa = 10.67QEE252 pKa = 3.96DD253 pKa = 4.33VLTDD257 pKa = 3.62VYY259 pKa = 11.53SDD261 pKa = 3.32AQEE264 pKa = 3.98YY265 pKa = 10.35QYY267 pKa = 11.52SQDD270 pKa = 3.04RR271 pKa = 11.84HH272 pKa = 5.75LSQVYY277 pKa = 9.21QLSQEE282 pKa = 4.31GEE284 pKa = 4.3SYY286 pKa = 10.99EE287 pKa = 4.78SNTHH291 pKa = 5.73GNTPIKK297 pKa = 10.68QEE299 pKa = 5.24DD300 pKa = 3.67DD301 pKa = 3.64DD302 pKa = 4.79NYY304 pKa = 11.5NGIAAQQYY312 pKa = 9.93GSTTFTTPCDD322 pKa = 3.6SGDD325 pKa = 4.26DD326 pKa = 4.01SDD328 pKa = 5.32HH329 pKa = 6.85SYY331 pKa = 11.13QGVEE335 pKa = 3.95YY336 pKa = 10.59LPNPVLATHH345 pKa = 7.41ILPGDD350 pKa = 3.68DD351 pKa = 3.72PEE353 pKa = 4.6FFEE356 pKa = 4.64GDD358 pKa = 3.54RR359 pKa = 11.84YY360 pKa = 11.13VLASPSLTEE369 pKa = 4.35HH370 pKa = 7.35DD371 pKa = 5.48LPGDD375 pKa = 4.01DD376 pKa = 6.02DD377 pKa = 5.52NDD379 pKa = 4.05FEE381 pKa = 6.29GDD383 pKa = 3.4QYY385 pKa = 11.86HH386 pKa = 7.21LARR389 pKa = 11.84PSFAEE394 pKa = 4.31NGLPDD399 pKa = 4.13NVEE402 pKa = 4.01KK403 pKa = 10.85DD404 pKa = 3.68DD405 pKa = 5.12EE406 pKa = 4.62NGQSHH411 pKa = 7.34LYY413 pKa = 9.76EE414 pKa = 4.66PPEE417 pKa = 4.23DD418 pKa = 3.72SNYY421 pKa = 10.57CEE423 pKa = 4.74DD424 pKa = 3.9QLGTLDD430 pKa = 4.38EE431 pKa = 4.49QNKK434 pKa = 9.86HH435 pKa = 6.47PDD437 pKa = 3.34TNINTDD443 pKa = 2.98TDD445 pKa = 3.51TDD447 pKa = 3.48TDD449 pKa = 3.9TNINITITINSDD461 pKa = 3.46SITNSNTNTMNVHH474 pKa = 5.57STDD477 pKa = 3.25KK478 pKa = 11.34AGDD481 pKa = 3.58GSEE484 pKa = 4.06GGGILEE490 pKa = 5.01EE491 pKa = 4.65NDD493 pKa = 3.86DD494 pKa = 4.0DD495 pKa = 5.9AGNQLEE501 pKa = 4.49EE502 pKa = 4.5SEE504 pKa = 4.93ADD506 pKa = 3.39HH507 pKa = 7.28DD508 pKa = 4.32HH509 pKa = 6.77GSVGTHH515 pKa = 5.44TPIIDD520 pKa = 3.95WNRR523 pKa = 11.84HH524 pKa = 4.78LAPSTAISDD533 pKa = 3.84PFSDD537 pKa = 4.67SEE539 pKa = 4.21IGHH542 pKa = 6.72DD543 pKa = 5.83LEE545 pKa = 7.27DD546 pKa = 4.16NDD548 pKa = 5.66DD549 pKa = 3.97NNEE552 pKa = 3.71FDD554 pKa = 4.98LFPAAHH560 pKa = 6.97EE561 pKa = 4.54DD562 pKa = 4.56DD563 pKa = 4.25YY564 pKa = 12.16AADD567 pKa = 3.7VNNMYY572 pKa = 7.76EE573 pKa = 4.4TTSPWNFDD581 pKa = 3.47DD582 pKa = 4.47ATEE585 pKa = 4.06FDD587 pKa = 3.96INQQYY592 pKa = 9.1TMVDD596 pKa = 4.24EE597 pKa = 4.51IAGDD601 pKa = 4.0DD602 pKa = 5.2DD603 pKa = 4.84IDD605 pKa = 4.28TPSEE609 pKa = 4.29MPTKK613 pKa = 10.73ADD615 pKa = 3.37DD616 pKa = 3.62TVTIDD621 pKa = 3.22THH623 pKa = 4.74TTAIDD628 pKa = 3.54NSSSSQAHH636 pKa = 5.85MDD638 pKa = 3.41STSVHH643 pKa = 6.6GISNQLEE650 pKa = 4.18SSPVDD655 pKa = 3.04THH657 pKa = 7.33LATEE661 pKa = 4.31AQNDD665 pKa = 3.9DD666 pKa = 4.08EE667 pKa = 5.12GADD670 pKa = 3.81DD671 pKa = 6.21LVDD674 pKa = 3.97EE675 pKa = 5.08YY676 pKa = 11.33EE677 pKa = 4.48DD678 pKa = 3.14VDD680 pKa = 3.87NRR682 pKa = 11.84VEE684 pKa = 3.97ASAEE688 pKa = 3.81ALEE691 pKa = 4.79NDD693 pKa = 4.5DD694 pKa = 4.99LTTTSPTLAGAHH706 pKa = 5.15VAGNDD711 pKa = 3.69ASDD714 pKa = 3.72SDD716 pKa = 4.01EE717 pKa = 4.65DD718 pKa = 5.97LIDD721 pKa = 3.94EE722 pKa = 5.41LEE724 pKa = 4.49NDD726 pKa = 4.46DD727 pKa = 5.98DD728 pKa = 6.63DD729 pKa = 6.4DD730 pKa = 6.01SEE732 pKa = 4.59YY733 pKa = 11.13QLDD736 pKa = 4.32PDD738 pKa = 4.35EE739 pKa = 5.27MDD741 pKa = 5.29DD742 pKa = 4.93SSSTNNSYY750 pKa = 10.71IEE752 pKa = 4.2TTGGEE757 pKa = 4.44GEE759 pKa = 4.29GRR761 pKa = 11.84NN762 pKa = 3.71
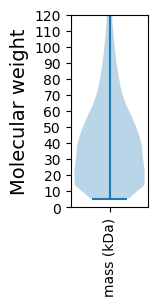
Molecular weight: 84.8 kDa
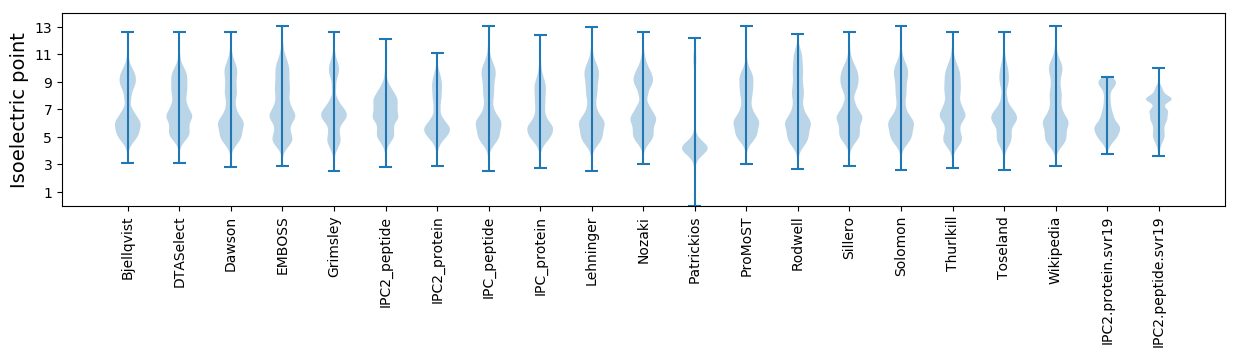
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X2ILU6|A0A1X2ILU6_9FUNG Mitochondrial glycine transporter OS=Absidia repens OX=90262 GN=BCR42DRAFT_411493 PE=3 SV=1
MM1 pKa = 7.62GNFTLGMLLVVMTMTRR17 pKa = 11.84RR18 pKa = 11.84LTMRR22 pKa = 11.84FTSRR26 pKa = 11.84LGRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84DD34 pKa = 3.01TTSIMGNFTLGMLLVIMTMTRR55 pKa = 11.84RR56 pKa = 11.84LTMRR60 pKa = 11.84FTSRR64 pKa = 11.84LGRR67 pKa = 11.84RR68 pKa = 11.84GRR70 pKa = 11.84DD71 pKa = 3.09TTSIMGNLTLSLATWFMVMGVFLLMMRR98 pKa = 11.84VTADD102 pKa = 3.07RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84NTLLLMLGVTSGIHH120 pKa = 5.84SSS122 pKa = 3.23
MM1 pKa = 7.62GNFTLGMLLVVMTMTRR17 pKa = 11.84RR18 pKa = 11.84LTMRR22 pKa = 11.84FTSRR26 pKa = 11.84LGRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84DD34 pKa = 3.01TTSIMGNFTLGMLLVIMTMTRR55 pKa = 11.84RR56 pKa = 11.84LTMRR60 pKa = 11.84FTSRR64 pKa = 11.84LGRR67 pKa = 11.84RR68 pKa = 11.84GRR70 pKa = 11.84DD71 pKa = 3.09TTSIMGNLTLSLATWFMVMGVFLLMMRR98 pKa = 11.84VTADD102 pKa = 3.07RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84NTLLLMLGVTSGIHH120 pKa = 5.84SSS122 pKa = 3.23
Molecular weight: 13.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6231592 |
49 |
7673 |
422.0 |
47.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.591 ± 0.018 | 1.387 ± 0.008 |
6.191 ± 0.017 | 5.196 ± 0.019 |
3.75 ± 0.014 | 5.371 ± 0.016 |
2.987 ± 0.012 | 5.486 ± 0.016 |
5.448 ± 0.02 | 9.168 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.606 ± 0.008 | 4.753 ± 0.012 |
5.122 ± 0.022 | 5.284 ± 0.022 |
4.891 ± 0.013 | 8.81 ± 0.031 |
6.777 ± 0.017 | 5.693 ± 0.02 |
1.196 ± 0.007 | 3.292 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
