
Torque teno virus 11
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 8.65
Get precalculated fractions of proteins
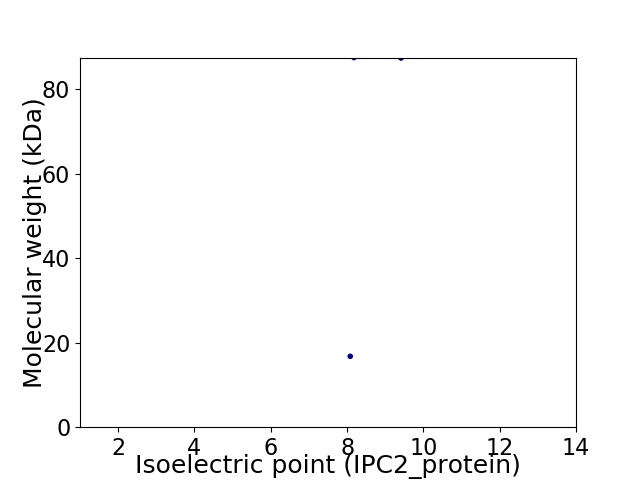
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q99AR0|Q99AR0_9VIRU Orf2 OS=Torque teno virus 11 OX=687350 PE=4 SV=1
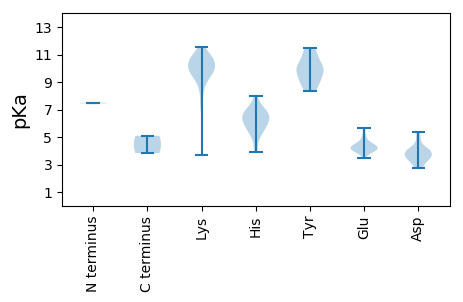
MM1 pKa = 7.5GKK3 pKa = 9.6CLKK6 pKa = 10.82KK7 pKa = 10.86NMFLGKK13 pKa = 9.75IYY15 pKa = 9.77RR16 pKa = 11.84QKK18 pKa = 10.71RR19 pKa = 11.84ALSLHH24 pKa = 5.68SVRR27 pKa = 11.84ASKK30 pKa = 10.43KK31 pKa = 9.62KK32 pKa = 9.81PPTAMSHH39 pKa = 5.21WSRR42 pKa = 11.84PVHH45 pKa = 5.86HH46 pKa = 6.91ATGIEE51 pKa = 4.07HH52 pKa = 7.45LWYY55 pKa = 10.11QSVINSHH62 pKa = 6.02SACCGCGDD70 pKa = 3.95PVRR73 pKa = 11.84HH74 pKa = 5.21FTNLAEE80 pKa = 4.7RR81 pKa = 11.84YY82 pKa = 9.39GFAPTPRR89 pKa = 11.84APPVAPTPTIRR100 pKa = 11.84RR101 pKa = 11.84ARR103 pKa = 11.84PLPVAPEE110 pKa = 3.73PRR112 pKa = 11.84ALPWHH117 pKa = 6.56GDD119 pKa = 3.08GGEE122 pKa = 3.92EE123 pKa = 4.19RR124 pKa = 11.84GTGGGDD130 pKa = 3.45GASPEE135 pKa = 4.72ADD137 pKa = 3.46FADD140 pKa = 5.31DD141 pKa = 4.85GLDD144 pKa = 3.75DD145 pKa = 5.35LVAALDD151 pKa = 3.82EE152 pKa = 4.41EE153 pKa = 4.72RR154 pKa = 5.09
MM1 pKa = 7.5GKK3 pKa = 9.6CLKK6 pKa = 10.82KK7 pKa = 10.86NMFLGKK13 pKa = 9.75IYY15 pKa = 9.77RR16 pKa = 11.84QKK18 pKa = 10.71RR19 pKa = 11.84ALSLHH24 pKa = 5.68SVRR27 pKa = 11.84ASKK30 pKa = 10.43KK31 pKa = 9.62KK32 pKa = 9.81PPTAMSHH39 pKa = 5.21WSRR42 pKa = 11.84PVHH45 pKa = 5.86HH46 pKa = 6.91ATGIEE51 pKa = 4.07HH52 pKa = 7.45LWYY55 pKa = 10.11QSVINSHH62 pKa = 6.02SACCGCGDD70 pKa = 3.95PVRR73 pKa = 11.84HH74 pKa = 5.21FTNLAEE80 pKa = 4.7RR81 pKa = 11.84YY82 pKa = 9.39GFAPTPRR89 pKa = 11.84APPVAPTPTIRR100 pKa = 11.84RR101 pKa = 11.84ARR103 pKa = 11.84PLPVAPEE110 pKa = 3.73PRR112 pKa = 11.84ALPWHH117 pKa = 6.56GDD119 pKa = 3.08GGEE122 pKa = 3.92EE123 pKa = 4.19RR124 pKa = 11.84GTGGGDD130 pKa = 3.45GASPEE135 pKa = 4.72ADD137 pKa = 3.46FADD140 pKa = 5.31DD141 pKa = 4.85GLDD144 pKa = 3.75DD145 pKa = 5.35LVAALDD151 pKa = 3.82EE152 pKa = 4.41EE153 pKa = 4.72RR154 pKa = 5.09
Molecular weight: 16.78 kDa
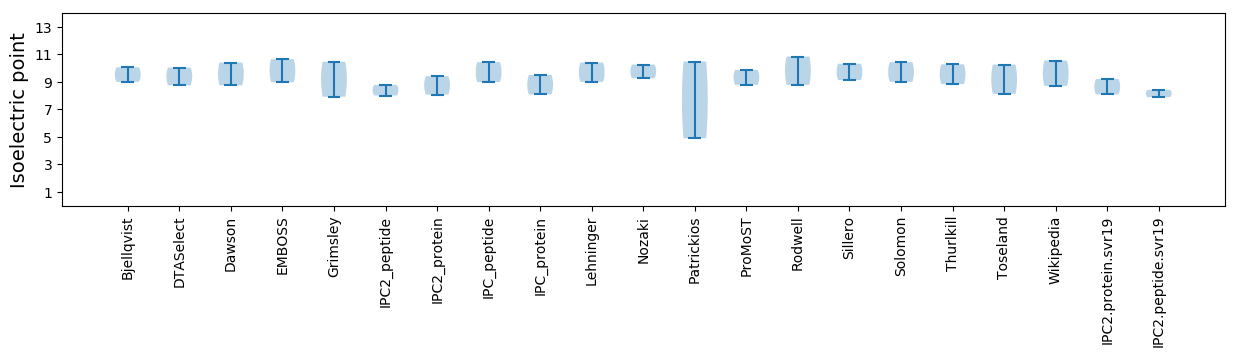
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q99AR0|Q99AR0_9VIRU Orf2 OS=Torque teno virus 11 OX=687350 PE=4 SV=1
MM1 pKa = 7.44AWGWWRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.5RR10 pKa = 11.84HH11 pKa = 3.92WWWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FARR20 pKa = 11.84SRR22 pKa = 11.84LRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84IRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84QWRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LFKK57 pKa = 10.16RR58 pKa = 11.84KK59 pKa = 8.74RR60 pKa = 11.84RR61 pKa = 11.84FWRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.94AKK70 pKa = 10.21IKK72 pKa = 8.44ITQWQPSRR80 pKa = 11.84VKK82 pKa = 8.42RR83 pKa = 11.84CFMIGYY89 pKa = 8.53FPAIICGQGRR99 pKa = 11.84WSEE102 pKa = 4.09NFTSHH107 pKa = 7.96IEE109 pKa = 4.11DD110 pKa = 3.96KK111 pKa = 10.54TPKK114 pKa = 10.02GPFGGGHH121 pKa = 4.3STARR125 pKa = 11.84LSLKK129 pKa = 10.26VLYY132 pKa = 10.32EE133 pKa = 4.27EE134 pKa = 4.43NQKK137 pKa = 9.32HH138 pKa = 5.87HH139 pKa = 6.34NFWTASNKK147 pKa = 9.06EE148 pKa = 3.91LEE150 pKa = 4.24LARR153 pKa = 11.84FFGSTWTLYY162 pKa = 9.92RR163 pKa = 11.84HH164 pKa = 6.04EE165 pKa = 4.47EE166 pKa = 3.49VDD168 pKa = 4.19YY169 pKa = 11.09ICTFNRR175 pKa = 11.84KK176 pKa = 9.08SPLGGNLLTGPSLHH190 pKa = 7.12PGAAMLSKK198 pKa = 10.65HH199 pKa = 6.3KK200 pKa = 10.6ILVPSFKK207 pKa = 9.81TRR209 pKa = 11.84PGGKK213 pKa = 7.38PTVKK217 pKa = 10.16FRR219 pKa = 11.84IKK221 pKa = 10.6PPTTLIDD228 pKa = 2.76KK229 pKa = 9.83WYY231 pKa = 8.45FQKK234 pKa = 10.74DD235 pKa = 3.17ICDD238 pKa = 3.65TTLLNINFVACDD250 pKa = 3.72LRR252 pKa = 11.84FPFCSPQTDD261 pKa = 3.77NVCVTFQVLSAVYY274 pKa = 10.68NDD276 pKa = 4.08FLSITNSKK284 pKa = 9.76LQATNWTDD292 pKa = 2.96EE293 pKa = 4.13YY294 pKa = 11.26TNFLNKK300 pKa = 10.02AFPSDD305 pKa = 3.39RR306 pKa = 11.84SINKK310 pKa = 9.53LNTFSTEE317 pKa = 3.99GAFSHH322 pKa = 6.65PKK324 pKa = 9.62VEE326 pKa = 4.12KK327 pKa = 10.14FKK329 pKa = 11.09PKK331 pKa = 10.05IPLPDD336 pKa = 3.38GEE338 pKa = 4.49YY339 pKa = 10.54FQEE342 pKa = 5.62KK343 pKa = 9.54DD344 pKa = 4.16ALWGDD349 pKa = 4.86PIHH352 pKa = 6.55ITTSQTRR359 pKa = 11.84QAIIQKK365 pKa = 9.33IVKK368 pKa = 9.77NATNYY373 pKa = 9.15LKK375 pKa = 10.65KK376 pKa = 10.21YY377 pKa = 8.32TDD379 pKa = 3.5EE380 pKa = 5.05FKK382 pKa = 10.82QFPNIAQFHH391 pKa = 6.65HH392 pKa = 6.8GFFTQWTGIYY402 pKa = 9.92SAAFLNYY409 pKa = 9.62HH410 pKa = 7.01RR411 pKa = 11.84ISPEE415 pKa = 3.84VPGLYY420 pKa = 10.04VDD422 pKa = 4.55TIYY425 pKa = 11.46NPLTDD430 pKa = 3.98KK431 pKa = 11.53GIGNHH436 pKa = 5.35IWIDD440 pKa = 3.9YY441 pKa = 10.12LSKK444 pKa = 10.87ADD446 pKa = 4.21PNLDD450 pKa = 3.54PVKK453 pKa = 10.05SKK455 pKa = 10.81CHH457 pKa = 6.15IADD460 pKa = 3.87LPLWMATFGLVDD472 pKa = 3.41WCKK475 pKa = 10.91KK476 pKa = 7.37EE477 pKa = 3.96TGNWGIPAWARR488 pKa = 11.84VLLICPYY495 pKa = 9.04TFPPMYY501 pKa = 9.98DD502 pKa = 2.85QKK504 pKa = 11.37NPNRR508 pKa = 11.84GFIPIGYY515 pKa = 8.78NFSQGLMPDD524 pKa = 3.48GSKK527 pKa = 10.6YY528 pKa = 10.69IPMQMRR534 pKa = 11.84AKK536 pKa = 9.87WYY538 pKa = 8.94PNLLNQQAVLEE549 pKa = 5.43DD550 pKa = 4.1ISQSGPFAFRR560 pKa = 11.84EE561 pKa = 4.32KK562 pKa = 10.53KK563 pKa = 9.95PSASITAKK571 pKa = 10.8YY572 pKa = 9.36KK573 pKa = 10.35FRR575 pKa = 11.84WHH577 pKa = 6.56FGGNPVSEE585 pKa = 4.11QVIADD590 pKa = 4.15PCQQPTFEE598 pKa = 4.43MPGTGSLPRR607 pKa = 11.84RR608 pKa = 11.84LQVTDD613 pKa = 3.91PKK615 pKa = 11.18LLGPHH620 pKa = 5.27YY621 pKa = 9.47TFHH624 pKa = 6.28SWDD627 pKa = 2.81WRR629 pKa = 11.84RR630 pKa = 11.84DD631 pKa = 3.43YY632 pKa = 11.31FNRR635 pKa = 11.84KK636 pKa = 3.72TLKK639 pKa = 10.47RR640 pKa = 11.84MSEE643 pKa = 3.94QQEE646 pKa = 4.01TSEE649 pKa = 4.47FLFPGPKK656 pKa = 9.43KK657 pKa = 10.2PRR659 pKa = 11.84VDD661 pKa = 3.72LGVVQAVEE669 pKa = 4.25KK670 pKa = 11.11VSFQAEE676 pKa = 4.18RR677 pKa = 11.84EE678 pKa = 4.14QKK680 pKa = 9.69PWEE683 pKa = 4.92DD684 pKa = 3.16SDD686 pKa = 4.18QPSEE690 pKa = 4.28TEE692 pKa = 3.76ALSEE696 pKa = 4.11EE697 pKa = 4.31EE698 pKa = 4.38EE699 pKa = 4.46EE700 pKa = 5.16GSIHH704 pKa = 5.77QQLRR708 pKa = 11.84QQLQQQKK715 pKa = 7.23QLRR718 pKa = 11.84RR719 pKa = 11.84GINCLFEE726 pKa = 4.5QLVKK730 pKa = 9.31TQQGVHH736 pKa = 6.95LDD738 pKa = 3.62PSLLL742 pKa = 3.83
MM1 pKa = 7.44AWGWWRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.5RR10 pKa = 11.84HH11 pKa = 3.92WWWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84FARR20 pKa = 11.84SRR22 pKa = 11.84LRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84IRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84TVRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84QWRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LFKK57 pKa = 10.16RR58 pKa = 11.84KK59 pKa = 8.74RR60 pKa = 11.84RR61 pKa = 11.84FWRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.94AKK70 pKa = 10.21IKK72 pKa = 8.44ITQWQPSRR80 pKa = 11.84VKK82 pKa = 8.42RR83 pKa = 11.84CFMIGYY89 pKa = 8.53FPAIICGQGRR99 pKa = 11.84WSEE102 pKa = 4.09NFTSHH107 pKa = 7.96IEE109 pKa = 4.11DD110 pKa = 3.96KK111 pKa = 10.54TPKK114 pKa = 10.02GPFGGGHH121 pKa = 4.3STARR125 pKa = 11.84LSLKK129 pKa = 10.26VLYY132 pKa = 10.32EE133 pKa = 4.27EE134 pKa = 4.43NQKK137 pKa = 9.32HH138 pKa = 5.87HH139 pKa = 6.34NFWTASNKK147 pKa = 9.06EE148 pKa = 3.91LEE150 pKa = 4.24LARR153 pKa = 11.84FFGSTWTLYY162 pKa = 9.92RR163 pKa = 11.84HH164 pKa = 6.04EE165 pKa = 4.47EE166 pKa = 3.49VDD168 pKa = 4.19YY169 pKa = 11.09ICTFNRR175 pKa = 11.84KK176 pKa = 9.08SPLGGNLLTGPSLHH190 pKa = 7.12PGAAMLSKK198 pKa = 10.65HH199 pKa = 6.3KK200 pKa = 10.6ILVPSFKK207 pKa = 9.81TRR209 pKa = 11.84PGGKK213 pKa = 7.38PTVKK217 pKa = 10.16FRR219 pKa = 11.84IKK221 pKa = 10.6PPTTLIDD228 pKa = 2.76KK229 pKa = 9.83WYY231 pKa = 8.45FQKK234 pKa = 10.74DD235 pKa = 3.17ICDD238 pKa = 3.65TTLLNINFVACDD250 pKa = 3.72LRR252 pKa = 11.84FPFCSPQTDD261 pKa = 3.77NVCVTFQVLSAVYY274 pKa = 10.68NDD276 pKa = 4.08FLSITNSKK284 pKa = 9.76LQATNWTDD292 pKa = 2.96EE293 pKa = 4.13YY294 pKa = 11.26TNFLNKK300 pKa = 10.02AFPSDD305 pKa = 3.39RR306 pKa = 11.84SINKK310 pKa = 9.53LNTFSTEE317 pKa = 3.99GAFSHH322 pKa = 6.65PKK324 pKa = 9.62VEE326 pKa = 4.12KK327 pKa = 10.14FKK329 pKa = 11.09PKK331 pKa = 10.05IPLPDD336 pKa = 3.38GEE338 pKa = 4.49YY339 pKa = 10.54FQEE342 pKa = 5.62KK343 pKa = 9.54DD344 pKa = 4.16ALWGDD349 pKa = 4.86PIHH352 pKa = 6.55ITTSQTRR359 pKa = 11.84QAIIQKK365 pKa = 9.33IVKK368 pKa = 9.77NATNYY373 pKa = 9.15LKK375 pKa = 10.65KK376 pKa = 10.21YY377 pKa = 8.32TDD379 pKa = 3.5EE380 pKa = 5.05FKK382 pKa = 10.82QFPNIAQFHH391 pKa = 6.65HH392 pKa = 6.8GFFTQWTGIYY402 pKa = 9.92SAAFLNYY409 pKa = 9.62HH410 pKa = 7.01RR411 pKa = 11.84ISPEE415 pKa = 3.84VPGLYY420 pKa = 10.04VDD422 pKa = 4.55TIYY425 pKa = 11.46NPLTDD430 pKa = 3.98KK431 pKa = 11.53GIGNHH436 pKa = 5.35IWIDD440 pKa = 3.9YY441 pKa = 10.12LSKK444 pKa = 10.87ADD446 pKa = 4.21PNLDD450 pKa = 3.54PVKK453 pKa = 10.05SKK455 pKa = 10.81CHH457 pKa = 6.15IADD460 pKa = 3.87LPLWMATFGLVDD472 pKa = 3.41WCKK475 pKa = 10.91KK476 pKa = 7.37EE477 pKa = 3.96TGNWGIPAWARR488 pKa = 11.84VLLICPYY495 pKa = 9.04TFPPMYY501 pKa = 9.98DD502 pKa = 2.85QKK504 pKa = 11.37NPNRR508 pKa = 11.84GFIPIGYY515 pKa = 8.78NFSQGLMPDD524 pKa = 3.48GSKK527 pKa = 10.6YY528 pKa = 10.69IPMQMRR534 pKa = 11.84AKK536 pKa = 9.87WYY538 pKa = 8.94PNLLNQQAVLEE549 pKa = 5.43DD550 pKa = 4.1ISQSGPFAFRR560 pKa = 11.84EE561 pKa = 4.32KK562 pKa = 10.53KK563 pKa = 9.95PSASITAKK571 pKa = 10.8YY572 pKa = 9.36KK573 pKa = 10.35FRR575 pKa = 11.84WHH577 pKa = 6.56FGGNPVSEE585 pKa = 4.11QVIADD590 pKa = 4.15PCQQPTFEE598 pKa = 4.43MPGTGSLPRR607 pKa = 11.84RR608 pKa = 11.84LQVTDD613 pKa = 3.91PKK615 pKa = 11.18LLGPHH620 pKa = 5.27YY621 pKa = 9.47TFHH624 pKa = 6.28SWDD627 pKa = 2.81WRR629 pKa = 11.84RR630 pKa = 11.84DD631 pKa = 3.43YY632 pKa = 11.31FNRR635 pKa = 11.84KK636 pKa = 3.72TLKK639 pKa = 10.47RR640 pKa = 11.84MSEE643 pKa = 3.94QQEE646 pKa = 4.01TSEE649 pKa = 4.47FLFPGPKK656 pKa = 9.43KK657 pKa = 10.2PRR659 pKa = 11.84VDD661 pKa = 3.72LGVVQAVEE669 pKa = 4.25KK670 pKa = 11.11VSFQAEE676 pKa = 4.18RR677 pKa = 11.84EE678 pKa = 4.14QKK680 pKa = 9.69PWEE683 pKa = 4.92DD684 pKa = 3.16SDD686 pKa = 4.18QPSEE690 pKa = 4.28TEE692 pKa = 3.76ALSEE696 pKa = 4.11EE697 pKa = 4.31EE698 pKa = 4.38EE699 pKa = 4.46EE700 pKa = 5.16GSIHH704 pKa = 5.77QQLRR708 pKa = 11.84QQLQQQKK715 pKa = 7.23QLRR718 pKa = 11.84RR719 pKa = 11.84GINCLFEE726 pKa = 4.5QLVKK730 pKa = 9.31TQQGVHH736 pKa = 6.95LDD738 pKa = 3.62PSLLL742 pKa = 3.83
Molecular weight: 87.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896 |
154 |
742 |
448.0 |
52.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.692 ± 2.452 | 1.786 ± 0.372 |
4.576 ± 0.581 | 4.799 ± 0.181 |
5.58 ± 1.368 | 6.362 ± 1.549 |
3.125 ± 0.949 | 4.799 ± 1.009 |
7.143 ± 0.893 | 7.478 ± 0.154 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.451 ± 0.228 | 3.906 ± 0.898 |
7.701 ± 1.233 | 5.022 ± 1.707 |
8.817 ± 0.172 | 5.692 ± 0.07 |
5.915 ± 0.628 | 4.018 ± 0.242 |
3.237 ± 0.591 | 2.902 ± 0.437 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
