
Tacheng Tick Virus 7
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
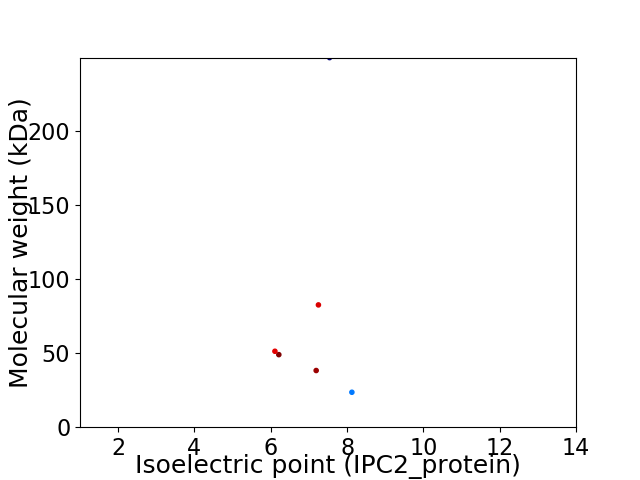
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXG9|A0A0B5KXG9_9RHAB GDP polyribonucleotidyltransferase OS=Tacheng Tick Virus 7 OX=1608089 GN=L PE=4 SV=1
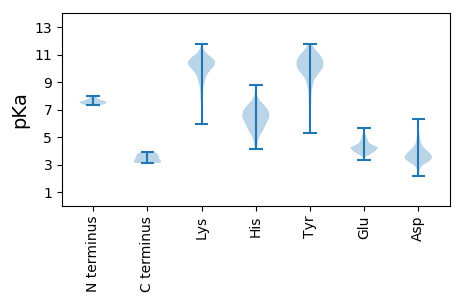
MM1 pKa = 7.99DD2 pKa = 5.87PSQKK6 pKa = 9.38ATQILGAFRR15 pKa = 11.84AGQADD20 pKa = 3.91TTVTATGEE28 pKa = 4.31SKK30 pKa = 10.05QWEE33 pKa = 4.68DD34 pKa = 4.14KK35 pKa = 10.8EE36 pKa = 4.69LDD38 pKa = 4.56AIPVTPLIALNEE50 pKa = 4.18AQLSQLFDD58 pKa = 3.69YY59 pKa = 11.11AVGEE63 pKa = 4.59LISPVPVNLPEE74 pKa = 4.49AVLLLASHH82 pKa = 6.05VTNPTSRR89 pKa = 11.84VDD91 pKa = 3.87DD92 pKa = 3.86NLLFILKK99 pKa = 9.8KK100 pKa = 10.0HH101 pKa = 5.87LLSKK105 pKa = 10.5EE106 pKa = 3.87LAPEE110 pKa = 4.19LNAIAEE116 pKa = 4.38KK117 pKa = 10.67LISTFKK123 pKa = 11.05AKK125 pKa = 10.07TGDD128 pKa = 3.42RR129 pKa = 11.84TTAAPPDD136 pKa = 3.58EE137 pKa = 4.47EE138 pKa = 3.97AVKK141 pKa = 10.51RR142 pKa = 11.84QEE144 pKa = 3.98QMLAAYY150 pKa = 8.63YY151 pKa = 9.65ISKK154 pKa = 8.49STAVTKK160 pKa = 10.75DD161 pKa = 3.02SLGPYY166 pKa = 7.66MKK168 pKa = 9.2WYY170 pKa = 10.49SDD172 pKa = 3.14ASVADD177 pKa = 3.88RR178 pKa = 11.84VEE180 pKa = 5.02FGCFMSFCLMRR191 pKa = 11.84SVASQEE197 pKa = 3.7VTIIRR202 pKa = 11.84NMGQEE207 pKa = 3.88IQPTGGSGGTTKK219 pKa = 10.72GGFYY223 pKa = 10.73GRR225 pKa = 11.84YY226 pKa = 7.62RR227 pKa = 11.84DD228 pKa = 3.88LYY230 pKa = 10.24RR231 pKa = 11.84GRR233 pKa = 11.84TFPSCGIQFVTQAAIAQIKK252 pKa = 9.61AAFSTGSQMLCSILVPVLRR271 pKa = 11.84VHH273 pKa = 6.65IQTDD277 pKa = 3.64PTPSVDD283 pKa = 3.27VQAVLRR289 pKa = 11.84YY290 pKa = 9.06GILMSLEE297 pKa = 3.77YY298 pKa = 10.3TGIYY302 pKa = 9.83LADD305 pKa = 3.66SYY307 pKa = 11.72LRR309 pKa = 11.84ALVRR313 pKa = 11.84TGEE316 pKa = 4.41TGSSLIPKK324 pKa = 8.95LYY326 pKa = 9.85WNVHH330 pKa = 4.63HH331 pKa = 7.32PEE333 pKa = 3.94YY334 pKa = 10.75EE335 pKa = 3.94RR336 pKa = 11.84LLDD339 pKa = 3.7MVQKK343 pKa = 10.58YY344 pKa = 7.62MRR346 pKa = 11.84DD347 pKa = 3.56NKK349 pKa = 10.33NARR352 pKa = 11.84AWRR355 pKa = 11.84WAKK358 pKa = 10.26CLHH361 pKa = 6.3SDD363 pKa = 3.67YY364 pKa = 11.19LSIYY368 pKa = 10.42SGVNNPWLCSLFTALFIDD386 pKa = 5.47KK387 pKa = 9.83EE388 pKa = 4.27PQPEE392 pKa = 4.33GTDD395 pKa = 3.12VWEE398 pKa = 4.4KK399 pKa = 10.18FCLRR403 pKa = 11.84KK404 pKa = 10.14SPEE407 pKa = 3.97HH408 pKa = 6.72KK409 pKa = 10.14EE410 pKa = 3.8WAQKK414 pKa = 9.94LAAAYY419 pKa = 8.13WAEE422 pKa = 4.16KK423 pKa = 9.62YY424 pKa = 10.3KK425 pKa = 11.15NSAITEE431 pKa = 4.16DD432 pKa = 3.41SDD434 pKa = 3.91LVKK437 pKa = 10.64RR438 pKa = 11.84IVGVAKK444 pKa = 9.88PVHH447 pKa = 6.68DD448 pKa = 5.19PYY450 pKa = 11.67DD451 pKa = 3.27VALRR455 pKa = 11.84MGSAEE460 pKa = 3.88
MM1 pKa = 7.99DD2 pKa = 5.87PSQKK6 pKa = 9.38ATQILGAFRR15 pKa = 11.84AGQADD20 pKa = 3.91TTVTATGEE28 pKa = 4.31SKK30 pKa = 10.05QWEE33 pKa = 4.68DD34 pKa = 4.14KK35 pKa = 10.8EE36 pKa = 4.69LDD38 pKa = 4.56AIPVTPLIALNEE50 pKa = 4.18AQLSQLFDD58 pKa = 3.69YY59 pKa = 11.11AVGEE63 pKa = 4.59LISPVPVNLPEE74 pKa = 4.49AVLLLASHH82 pKa = 6.05VTNPTSRR89 pKa = 11.84VDD91 pKa = 3.87DD92 pKa = 3.86NLLFILKK99 pKa = 9.8KK100 pKa = 10.0HH101 pKa = 5.87LLSKK105 pKa = 10.5EE106 pKa = 3.87LAPEE110 pKa = 4.19LNAIAEE116 pKa = 4.38KK117 pKa = 10.67LISTFKK123 pKa = 11.05AKK125 pKa = 10.07TGDD128 pKa = 3.42RR129 pKa = 11.84TTAAPPDD136 pKa = 3.58EE137 pKa = 4.47EE138 pKa = 3.97AVKK141 pKa = 10.51RR142 pKa = 11.84QEE144 pKa = 3.98QMLAAYY150 pKa = 8.63YY151 pKa = 9.65ISKK154 pKa = 8.49STAVTKK160 pKa = 10.75DD161 pKa = 3.02SLGPYY166 pKa = 7.66MKK168 pKa = 9.2WYY170 pKa = 10.49SDD172 pKa = 3.14ASVADD177 pKa = 3.88RR178 pKa = 11.84VEE180 pKa = 5.02FGCFMSFCLMRR191 pKa = 11.84SVASQEE197 pKa = 3.7VTIIRR202 pKa = 11.84NMGQEE207 pKa = 3.88IQPTGGSGGTTKK219 pKa = 10.72GGFYY223 pKa = 10.73GRR225 pKa = 11.84YY226 pKa = 7.62RR227 pKa = 11.84DD228 pKa = 3.88LYY230 pKa = 10.24RR231 pKa = 11.84GRR233 pKa = 11.84TFPSCGIQFVTQAAIAQIKK252 pKa = 9.61AAFSTGSQMLCSILVPVLRR271 pKa = 11.84VHH273 pKa = 6.65IQTDD277 pKa = 3.64PTPSVDD283 pKa = 3.27VQAVLRR289 pKa = 11.84YY290 pKa = 9.06GILMSLEE297 pKa = 3.77YY298 pKa = 10.3TGIYY302 pKa = 9.83LADD305 pKa = 3.66SYY307 pKa = 11.72LRR309 pKa = 11.84ALVRR313 pKa = 11.84TGEE316 pKa = 4.41TGSSLIPKK324 pKa = 8.95LYY326 pKa = 9.85WNVHH330 pKa = 4.63HH331 pKa = 7.32PEE333 pKa = 3.94YY334 pKa = 10.75EE335 pKa = 3.94RR336 pKa = 11.84LLDD339 pKa = 3.7MVQKK343 pKa = 10.58YY344 pKa = 7.62MRR346 pKa = 11.84DD347 pKa = 3.56NKK349 pKa = 10.33NARR352 pKa = 11.84AWRR355 pKa = 11.84WAKK358 pKa = 10.26CLHH361 pKa = 6.3SDD363 pKa = 3.67YY364 pKa = 11.19LSIYY368 pKa = 10.42SGVNNPWLCSLFTALFIDD386 pKa = 5.47KK387 pKa = 9.83EE388 pKa = 4.27PQPEE392 pKa = 4.33GTDD395 pKa = 3.12VWEE398 pKa = 4.4KK399 pKa = 10.18FCLRR403 pKa = 11.84KK404 pKa = 10.14SPEE407 pKa = 3.97HH408 pKa = 6.72KK409 pKa = 10.14EE410 pKa = 3.8WAQKK414 pKa = 9.94LAAAYY419 pKa = 8.13WAEE422 pKa = 4.16KK423 pKa = 9.62YY424 pKa = 10.3KK425 pKa = 11.15NSAITEE431 pKa = 4.16DD432 pKa = 3.41SDD434 pKa = 3.91LVKK437 pKa = 10.64RR438 pKa = 11.84IVGVAKK444 pKa = 9.88PVHH447 pKa = 6.68DD448 pKa = 5.19PYY450 pKa = 11.67DD451 pKa = 3.27VALRR455 pKa = 11.84MGSAEE460 pKa = 3.88
Molecular weight: 51.27 kDa
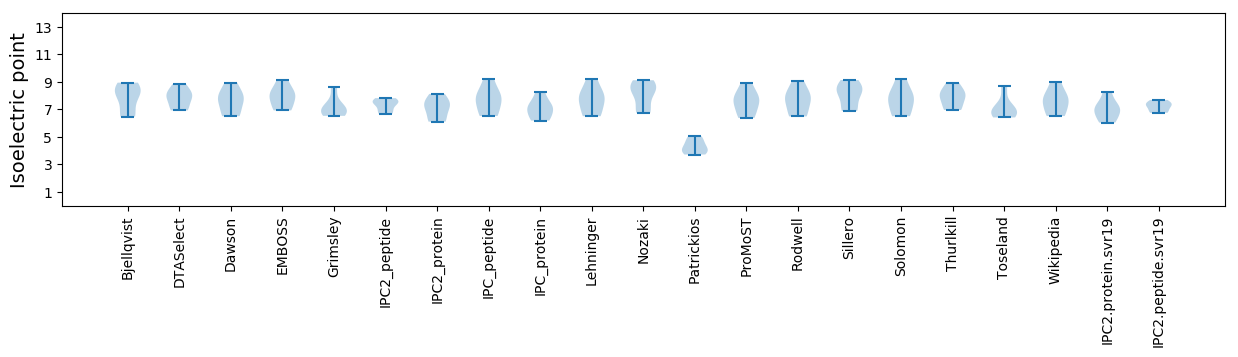
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRJ9|A0A0B5KRJ9_9RHAB ORF4 OS=Tacheng Tick Virus 7 OX=1608089 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.48SKK3 pKa = 10.23HH4 pKa = 5.98PKK6 pKa = 9.93CNPTLYY12 pKa = 10.74LSIEE16 pKa = 4.41GKK18 pKa = 10.33VEE20 pKa = 3.54VHH22 pKa = 5.27TLVRR26 pKa = 11.84PVLDD30 pKa = 4.32KK31 pKa = 10.98IVGRR35 pKa = 11.84LLITSGCPSSGDD47 pKa = 3.47LKK49 pKa = 10.25HH50 pKa = 6.4HH51 pKa = 5.58VRR53 pKa = 11.84AAIAGIIHH61 pKa = 7.06MLTFDD66 pKa = 3.99PVRR69 pKa = 11.84GRR71 pKa = 11.84HH72 pKa = 6.3LLMTDD77 pKa = 3.53GGPRR81 pKa = 11.84MGRR84 pKa = 11.84QLIWNMSASLIYY96 pKa = 10.88KK97 pKa = 6.33MTRR100 pKa = 11.84DD101 pKa = 3.52PRR103 pKa = 11.84VAQLPTFEE111 pKa = 4.47WSPHH115 pKa = 4.1GTFSSEE121 pKa = 4.22DD122 pKa = 3.36GTVTKK127 pKa = 9.67WQVHH131 pKa = 5.05VKK133 pKa = 9.64TSTAPPSLAALNIPPGRR150 pKa = 11.84ITDD153 pKa = 3.66QEE155 pKa = 4.18YY156 pKa = 8.71MRR158 pKa = 11.84ARR160 pKa = 11.84SLVARR165 pKa = 11.84FHH167 pKa = 6.39PPEE170 pKa = 4.52GSRR173 pKa = 11.84EE174 pKa = 4.06PTSPRR179 pKa = 11.84GDD181 pKa = 3.26TSGSSQGEE189 pKa = 4.38EE190 pKa = 4.01EE191 pKa = 3.92EE192 pKa = 4.31HH193 pKa = 7.66LYY195 pKa = 10.27EE196 pKa = 4.8EE197 pKa = 4.87PMDD200 pKa = 4.1FLTDD204 pKa = 4.23LGGLKK209 pKa = 9.21QRR211 pKa = 11.84RR212 pKa = 3.68
MM1 pKa = 7.48SKK3 pKa = 10.23HH4 pKa = 5.98PKK6 pKa = 9.93CNPTLYY12 pKa = 10.74LSIEE16 pKa = 4.41GKK18 pKa = 10.33VEE20 pKa = 3.54VHH22 pKa = 5.27TLVRR26 pKa = 11.84PVLDD30 pKa = 4.32KK31 pKa = 10.98IVGRR35 pKa = 11.84LLITSGCPSSGDD47 pKa = 3.47LKK49 pKa = 10.25HH50 pKa = 6.4HH51 pKa = 5.58VRR53 pKa = 11.84AAIAGIIHH61 pKa = 7.06MLTFDD66 pKa = 3.99PVRR69 pKa = 11.84GRR71 pKa = 11.84HH72 pKa = 6.3LLMTDD77 pKa = 3.53GGPRR81 pKa = 11.84MGRR84 pKa = 11.84QLIWNMSASLIYY96 pKa = 10.88KK97 pKa = 6.33MTRR100 pKa = 11.84DD101 pKa = 3.52PRR103 pKa = 11.84VAQLPTFEE111 pKa = 4.47WSPHH115 pKa = 4.1GTFSSEE121 pKa = 4.22DD122 pKa = 3.36GTVTKK127 pKa = 9.67WQVHH131 pKa = 5.05VKK133 pKa = 9.64TSTAPPSLAALNIPPGRR150 pKa = 11.84ITDD153 pKa = 3.66QEE155 pKa = 4.18YY156 pKa = 8.71MRR158 pKa = 11.84ARR160 pKa = 11.84SLVARR165 pKa = 11.84FHH167 pKa = 6.39PPEE170 pKa = 4.52GSRR173 pKa = 11.84EE174 pKa = 4.06PTSPRR179 pKa = 11.84GDD181 pKa = 3.26TSGSSQGEE189 pKa = 4.38EE190 pKa = 4.01EE191 pKa = 3.92EE192 pKa = 4.31HH193 pKa = 7.66LYY195 pKa = 10.27EE196 pKa = 4.8EE197 pKa = 4.87PMDD200 pKa = 4.1FLTDD204 pKa = 4.23LGGLKK209 pKa = 9.21QRR211 pKa = 11.84RR212 pKa = 3.68
Molecular weight: 23.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4409 |
212 |
2215 |
734.8 |
82.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.555 ± 0.431 | 1.86 ± 0.249 |
4.854 ± 0.449 | 4.922 ± 0.381 |
3.357 ± 0.157 | 5.648 ± 0.213 |
3.402 ± 0.401 | 5.466 ± 0.361 |
4.967 ± 0.365 | 11.272 ± 0.553 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.949 ± 0.136 | 3.266 ± 0.169 |
6.056 ± 0.691 | 3.946 ± 0.331 |
5.262 ± 0.282 | 8.868 ± 0.515 |
6.305 ± 0.099 | 6.033 ± 0.319 |
1.814 ± 0.039 | 3.198 ± 0.329 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
