
Trifolium-associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
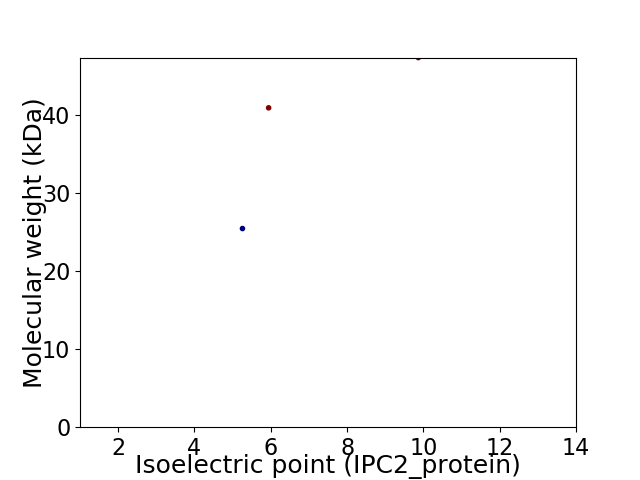
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4U8W1|A0A0B4U8W1_9VIRU Uncharacterized protein OS=Trifolium-associated circular DNA virus 1 OX=1590173 PE=4 SV=1
MM1 pKa = 7.84ARR3 pKa = 11.84QQHH6 pKa = 4.52QLEE9 pKa = 4.42RR10 pKa = 11.84SADD13 pKa = 3.68LQRR16 pKa = 11.84TNGTTTGSSQHH27 pKa = 6.66DD28 pKa = 3.95EE29 pKa = 4.09VRR31 pKa = 11.84GEE33 pKa = 3.98QTALRR38 pKa = 11.84SNDD41 pKa = 3.58SQHH44 pKa = 6.12VEE46 pKa = 3.91PGRR49 pKa = 11.84GHH51 pKa = 6.23NALRR55 pKa = 11.84LCDD58 pKa = 3.85EE59 pKa = 4.54VGCRR63 pKa = 11.84PEE65 pKa = 4.44PSTATEE71 pKa = 4.47PNPQSGGPAGGLEE84 pKa = 3.99EE85 pKa = 4.44RR86 pKa = 11.84SGGGEE91 pKa = 4.0DD92 pKa = 3.74SRR94 pKa = 11.84GRR96 pKa = 11.84LPGSDD101 pKa = 3.75YY102 pKa = 11.23DD103 pKa = 3.6QQRR106 pKa = 11.84WHH108 pKa = 6.91HH109 pKa = 6.1PVPLVSILQALQSCEE124 pKa = 3.77GHH126 pKa = 5.93SQDD129 pKa = 4.0TLVGGSTPPPCYY141 pKa = 10.1SPPSEE146 pKa = 4.17HH147 pKa = 6.22VRR149 pKa = 11.84HH150 pKa = 6.15TGGQDD155 pKa = 3.25RR156 pKa = 11.84EE157 pKa = 4.07HH158 pKa = 6.73SRR160 pKa = 11.84YY161 pKa = 9.87YY162 pKa = 11.02ADD164 pKa = 3.13QLEE167 pKa = 4.56YY168 pKa = 10.79QRR170 pKa = 11.84DD171 pKa = 3.17IHH173 pKa = 6.11PWINGLYY180 pKa = 8.83DD181 pKa = 3.33TGSQWEE187 pKa = 4.01YY188 pKa = 9.6RR189 pKa = 11.84QQSGRR194 pKa = 11.84GDD196 pKa = 3.59QQSDD200 pKa = 3.73RR201 pKa = 11.84DD202 pKa = 3.81DD203 pKa = 4.02SSCFGCSYY211 pKa = 11.38KK212 pKa = 10.57DD213 pKa = 3.08ICILFSVHH221 pKa = 6.33SGASPTRR228 pKa = 11.84QLL230 pKa = 3.75
MM1 pKa = 7.84ARR3 pKa = 11.84QQHH6 pKa = 4.52QLEE9 pKa = 4.42RR10 pKa = 11.84SADD13 pKa = 3.68LQRR16 pKa = 11.84TNGTTTGSSQHH27 pKa = 6.66DD28 pKa = 3.95EE29 pKa = 4.09VRR31 pKa = 11.84GEE33 pKa = 3.98QTALRR38 pKa = 11.84SNDD41 pKa = 3.58SQHH44 pKa = 6.12VEE46 pKa = 3.91PGRR49 pKa = 11.84GHH51 pKa = 6.23NALRR55 pKa = 11.84LCDD58 pKa = 3.85EE59 pKa = 4.54VGCRR63 pKa = 11.84PEE65 pKa = 4.44PSTATEE71 pKa = 4.47PNPQSGGPAGGLEE84 pKa = 3.99EE85 pKa = 4.44RR86 pKa = 11.84SGGGEE91 pKa = 4.0DD92 pKa = 3.74SRR94 pKa = 11.84GRR96 pKa = 11.84LPGSDD101 pKa = 3.75YY102 pKa = 11.23DD103 pKa = 3.6QQRR106 pKa = 11.84WHH108 pKa = 6.91HH109 pKa = 6.1PVPLVSILQALQSCEE124 pKa = 3.77GHH126 pKa = 5.93SQDD129 pKa = 4.0TLVGGSTPPPCYY141 pKa = 10.1SPPSEE146 pKa = 4.17HH147 pKa = 6.22VRR149 pKa = 11.84HH150 pKa = 6.15TGGQDD155 pKa = 3.25RR156 pKa = 11.84EE157 pKa = 4.07HH158 pKa = 6.73SRR160 pKa = 11.84YY161 pKa = 9.87YY162 pKa = 11.02ADD164 pKa = 3.13QLEE167 pKa = 4.56YY168 pKa = 10.79QRR170 pKa = 11.84DD171 pKa = 3.17IHH173 pKa = 6.11PWINGLYY180 pKa = 8.83DD181 pKa = 3.33TGSQWEE187 pKa = 4.01YY188 pKa = 9.6RR189 pKa = 11.84QQSGRR194 pKa = 11.84GDD196 pKa = 3.59QQSDD200 pKa = 3.73RR201 pKa = 11.84DD202 pKa = 3.81DD203 pKa = 4.02SSCFGCSYY211 pKa = 11.38KK212 pKa = 10.57DD213 pKa = 3.08ICILFSVHH221 pKa = 6.33SGASPTRR228 pKa = 11.84QLL230 pKa = 3.75
Molecular weight: 25.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4U8Q6|A0A0B4U8Q6_9VIRU Replication-associated protein OS=Trifolium-associated circular DNA virus 1 OX=1590173 PE=3 SV=1
MM1 pKa = 7.78AYY3 pKa = 9.81KK4 pKa = 10.62LAGRR8 pKa = 11.84AAGATLGFIYY18 pKa = 10.69NNTKK22 pKa = 10.2GALIGARR29 pKa = 11.84MAKK32 pKa = 9.46YY33 pKa = 10.16ARR35 pKa = 11.84TGIRR39 pKa = 11.84ARR41 pKa = 11.84VARR44 pKa = 11.84SNKK47 pKa = 9.58RR48 pKa = 11.84KK49 pKa = 7.66ATVYY53 pKa = 10.64AGTKK57 pKa = 9.68RR58 pKa = 11.84RR59 pKa = 11.84GSVLAAPPAKK69 pKa = 9.85RR70 pKa = 11.84RR71 pKa = 11.84VFKK74 pKa = 10.83SRR76 pKa = 11.84PRR78 pKa = 11.84KK79 pKa = 7.81QLRR82 pKa = 11.84GGKK85 pKa = 9.25RR86 pKa = 11.84RR87 pKa = 11.84GGGSKK92 pKa = 10.27RR93 pKa = 11.84SSFANAIAKK102 pKa = 8.73TGASNFSIGSGAKK115 pKa = 9.69LYY117 pKa = 11.03SGFKK121 pKa = 9.24RR122 pKa = 11.84TQLPTTFNCTVTQRR136 pKa = 11.84VSGTSSTQSVVLLPTYY152 pKa = 10.11GLRR155 pKa = 11.84ATGEE159 pKa = 4.24TGDD162 pKa = 4.27TASQSQFTIDD172 pKa = 4.83KK173 pKa = 10.26SLLDD177 pKa = 3.58TAEE180 pKa = 3.73RR181 pKa = 11.84WLANNTNSSAAQTYY195 pKa = 9.99SGPTGRR201 pKa = 11.84RR202 pKa = 11.84QGVRR206 pKa = 11.84NTMKK210 pKa = 10.32FVVNKK215 pKa = 9.72LHH217 pKa = 6.26YY218 pKa = 10.5DD219 pKa = 3.33QMIRR223 pKa = 11.84NMSNQDD229 pKa = 3.18ADD231 pKa = 3.33ITLYY235 pKa = 10.97DD236 pKa = 3.56CVMRR240 pKa = 11.84SGVDD244 pKa = 3.39PNQAPQLSPIPNQADD259 pKa = 3.6PLADD263 pKa = 3.01WRR265 pKa = 11.84NGLEE269 pKa = 4.06VEE271 pKa = 4.91RR272 pKa = 11.84IAAGAYY278 pKa = 10.2LGATTINSVGTTPFHH293 pKa = 7.44SSAFCKK299 pKa = 10.22LYY301 pKa = 10.52RR302 pKa = 11.84VAKK305 pKa = 7.6VTRR308 pKa = 11.84KK309 pKa = 7.32TLSSGEE315 pKa = 4.18VHH317 pKa = 6.01HH318 pKa = 6.53HH319 pKa = 6.04HH320 pKa = 5.78VTVRR324 pKa = 11.84PRR326 pKa = 11.84NMFDD330 pKa = 3.26TQVDD334 pKa = 4.12KK335 pKa = 11.09IGNIPDD341 pKa = 3.29ITQTNLNTRR350 pKa = 11.84GTYY353 pKa = 9.76IPGLTGFTILVASGSIGNNPDD374 pKa = 2.85VATNNQIGMTAVALDD389 pKa = 3.91VVTKK393 pKa = 9.43TSASFSQFTRR403 pKa = 11.84EE404 pKa = 3.8RR405 pKa = 11.84RR406 pKa = 11.84QHH408 pKa = 5.76VNFDD412 pKa = 3.48GMSTNNITAVVLDD425 pKa = 4.24DD426 pKa = 3.67TDD428 pKa = 6.13AITTVQAAGGGVV440 pKa = 3.02
MM1 pKa = 7.78AYY3 pKa = 9.81KK4 pKa = 10.62LAGRR8 pKa = 11.84AAGATLGFIYY18 pKa = 10.69NNTKK22 pKa = 10.2GALIGARR29 pKa = 11.84MAKK32 pKa = 9.46YY33 pKa = 10.16ARR35 pKa = 11.84TGIRR39 pKa = 11.84ARR41 pKa = 11.84VARR44 pKa = 11.84SNKK47 pKa = 9.58RR48 pKa = 11.84KK49 pKa = 7.66ATVYY53 pKa = 10.64AGTKK57 pKa = 9.68RR58 pKa = 11.84RR59 pKa = 11.84GSVLAAPPAKK69 pKa = 9.85RR70 pKa = 11.84RR71 pKa = 11.84VFKK74 pKa = 10.83SRR76 pKa = 11.84PRR78 pKa = 11.84KK79 pKa = 7.81QLRR82 pKa = 11.84GGKK85 pKa = 9.25RR86 pKa = 11.84RR87 pKa = 11.84GGGSKK92 pKa = 10.27RR93 pKa = 11.84SSFANAIAKK102 pKa = 8.73TGASNFSIGSGAKK115 pKa = 9.69LYY117 pKa = 11.03SGFKK121 pKa = 9.24RR122 pKa = 11.84TQLPTTFNCTVTQRR136 pKa = 11.84VSGTSSTQSVVLLPTYY152 pKa = 10.11GLRR155 pKa = 11.84ATGEE159 pKa = 4.24TGDD162 pKa = 4.27TASQSQFTIDD172 pKa = 4.83KK173 pKa = 10.26SLLDD177 pKa = 3.58TAEE180 pKa = 3.73RR181 pKa = 11.84WLANNTNSSAAQTYY195 pKa = 9.99SGPTGRR201 pKa = 11.84RR202 pKa = 11.84QGVRR206 pKa = 11.84NTMKK210 pKa = 10.32FVVNKK215 pKa = 9.72LHH217 pKa = 6.26YY218 pKa = 10.5DD219 pKa = 3.33QMIRR223 pKa = 11.84NMSNQDD229 pKa = 3.18ADD231 pKa = 3.33ITLYY235 pKa = 10.97DD236 pKa = 3.56CVMRR240 pKa = 11.84SGVDD244 pKa = 3.39PNQAPQLSPIPNQADD259 pKa = 3.6PLADD263 pKa = 3.01WRR265 pKa = 11.84NGLEE269 pKa = 4.06VEE271 pKa = 4.91RR272 pKa = 11.84IAAGAYY278 pKa = 10.2LGATTINSVGTTPFHH293 pKa = 7.44SSAFCKK299 pKa = 10.22LYY301 pKa = 10.52RR302 pKa = 11.84VAKK305 pKa = 7.6VTRR308 pKa = 11.84KK309 pKa = 7.32TLSSGEE315 pKa = 4.18VHH317 pKa = 6.01HH318 pKa = 6.53HH319 pKa = 6.04HH320 pKa = 5.78VTVRR324 pKa = 11.84PRR326 pKa = 11.84NMFDD330 pKa = 3.26TQVDD334 pKa = 4.12KK335 pKa = 11.09IGNIPDD341 pKa = 3.29ITQTNLNTRR350 pKa = 11.84GTYY353 pKa = 9.76IPGLTGFTILVASGSIGNNPDD374 pKa = 2.85VATNNQIGMTAVALDD389 pKa = 3.91VVTKK393 pKa = 9.43TSASFSQFTRR403 pKa = 11.84EE404 pKa = 3.8RR405 pKa = 11.84RR406 pKa = 11.84QHH408 pKa = 5.76VNFDD412 pKa = 3.48GMSTNNITAVVLDD425 pKa = 4.24DD426 pKa = 3.67TDD428 pKa = 6.13AITTVQAAGGGVV440 pKa = 3.02
Molecular weight: 47.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1025 |
230 |
440 |
341.7 |
37.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.902 ± 1.453 | 2.146 ± 0.808 |
6.146 ± 1.001 | 3.902 ± 1.368 |
3.61 ± 0.949 | 8.098 ± 1.561 |
3.122 ± 0.845 | 4.39 ± 0.667 |
4.39 ± 1.125 | 6.341 ± 0.246 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.366 ± 0.375 | 5.171 ± 0.868 |
4.78 ± 0.702 | 5.073 ± 1.269 |
7.415 ± 0.774 | 7.415 ± 1.357 |
7.902 ± 1.622 | 6.341 ± 0.792 |
1.561 ± 0.737 | 2.927 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
