
Hubei permutotetra-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.22
Get precalculated fractions of proteins
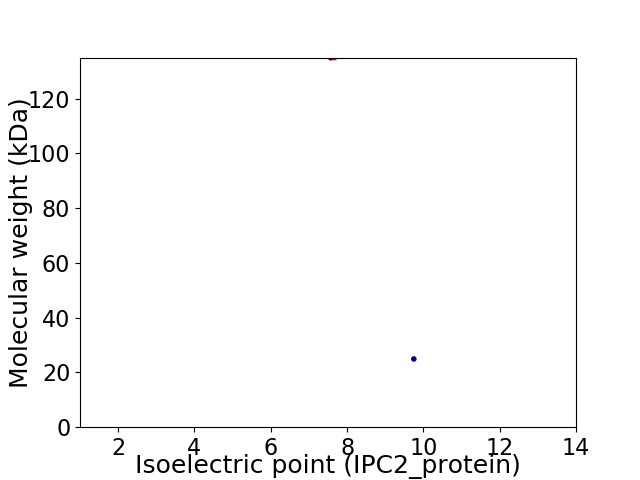
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHW9|A0A1L3KHW9_9VIRU RdRp OS=Hubei permutotetra-like virus 7 OX=1923081 PE=4 SV=1
MM1 pKa = 7.89DD2 pKa = 5.69ASNPIIPNTLTTMSNILDD20 pKa = 3.53RR21 pKa = 11.84QARR24 pKa = 11.84MTRR27 pKa = 11.84QDD29 pKa = 3.35FSLLRR34 pKa = 11.84DD35 pKa = 3.58RR36 pKa = 11.84LSTQARR42 pKa = 11.84HH43 pKa = 5.57LLEE46 pKa = 4.75VDD48 pKa = 3.83VPRR51 pKa = 11.84LPPEE55 pKa = 3.83QVQEE59 pKa = 3.73ILKK62 pKa = 9.44EE63 pKa = 4.09YY64 pKa = 10.63KK65 pKa = 7.67EE66 pKa = 4.37TRR68 pKa = 11.84VAVPPDD74 pKa = 3.37SKK76 pKa = 11.59APLEE80 pKa = 4.36FPTLDD85 pKa = 4.09LYY87 pKa = 11.47AEE89 pKa = 5.29EE90 pKa = 4.61IPISRR95 pKa = 11.84KK96 pKa = 9.75LDD98 pKa = 3.38FDD100 pKa = 3.6GRR102 pKa = 11.84PIHH105 pKa = 6.66PAGTNSKK112 pKa = 9.75QGRR115 pKa = 11.84VHH117 pKa = 6.85LSKK120 pKa = 11.08SFGSQPRR127 pKa = 11.84PMEE130 pKa = 4.3GVSYY134 pKa = 10.25LAEE137 pKa = 3.77ILGRR141 pKa = 11.84DD142 pKa = 3.7VCNTTHH148 pKa = 7.44IYY150 pKa = 10.29TSGTRR155 pKa = 11.84AGFQKK160 pKa = 10.25RR161 pKa = 11.84VAKK164 pKa = 10.1QLGRR168 pKa = 11.84KK169 pKa = 8.76AVSLAKK175 pKa = 9.69MFRR178 pKa = 11.84EE179 pKa = 4.07VRR181 pKa = 11.84GLGKK185 pKa = 10.23NPTAKK190 pKa = 10.33YY191 pKa = 9.93VNAYY195 pKa = 10.33LDD197 pKa = 3.43TLMPRR202 pKa = 11.84KK203 pKa = 9.67PNTNWPGVHH212 pKa = 6.75TDD214 pKa = 4.63LIDD217 pKa = 5.14GIMDD221 pKa = 5.3GIKK224 pKa = 10.29INFKK228 pKa = 10.55SSAGAPYY235 pKa = 9.91HH236 pKa = 7.12RR237 pKa = 11.84PLHH240 pKa = 6.03LCADD244 pKa = 4.16MVLSTGLPAAIEE256 pKa = 4.24IIKK259 pKa = 10.23NSKK262 pKa = 9.14PIHH265 pKa = 6.73DD266 pKa = 4.1AMEE269 pKa = 5.36DD270 pKa = 3.48DD271 pKa = 4.1VEE273 pKa = 4.46FFISEE278 pKa = 4.58LKK280 pKa = 10.99NKK282 pKa = 7.79TDD284 pKa = 3.35RR285 pKa = 11.84YY286 pKa = 10.56DD287 pKa = 3.42VNEE290 pKa = 4.51LMDD293 pKa = 3.4KK294 pKa = 9.52TRR296 pKa = 11.84PYY298 pKa = 10.81CCLPAHH304 pKa = 6.3WRR306 pKa = 11.84LLFSLVSQGFSATHH320 pKa = 5.26PTFDD324 pKa = 4.51EE325 pKa = 4.25IKK327 pKa = 9.43SANAYY332 pKa = 8.77GHH334 pKa = 6.41SNAHH338 pKa = 5.14GGMRR342 pKa = 11.84RR343 pKa = 11.84RR344 pKa = 11.84YY345 pKa = 8.92DD346 pKa = 2.87WAMTANTRR354 pKa = 11.84GQVIAYY360 pKa = 9.67GDD362 pKa = 3.78DD363 pKa = 3.2ALISIKK369 pKa = 10.35RR370 pKa = 11.84GSDD373 pKa = 2.9IFLVAPDD380 pKa = 4.13FRR382 pKa = 11.84QMDD385 pKa = 4.11GSIHH389 pKa = 6.85RR390 pKa = 11.84EE391 pKa = 3.7DD392 pKa = 2.89IKK394 pKa = 11.54YY395 pKa = 8.86IFRR398 pKa = 11.84WILSHH403 pKa = 7.52LIRR406 pKa = 11.84DD407 pKa = 3.89APGADD412 pKa = 3.33VNFWGPLFSLWEE424 pKa = 4.05FFAIDD429 pKa = 4.92PILMCEE435 pKa = 4.01GSKK438 pKa = 10.72AYY440 pKa = 8.69TKK442 pKa = 10.54KK443 pKa = 10.8NPDD446 pKa = 3.17AGMLSGVPGTTLINTVKK463 pKa = 10.7CATAWDD469 pKa = 4.92LYY471 pKa = 11.19LEE473 pKa = 4.69HH474 pKa = 7.19CANTGSDD481 pKa = 3.74ALDD484 pKa = 3.98PDD486 pKa = 3.52MARR489 pKa = 11.84RR490 pKa = 11.84FMAKK494 pKa = 9.75HH495 pKa = 5.24GLEE498 pKa = 4.2LKK500 pKa = 10.34PGTFSPQRR508 pKa = 11.84LPTPDD513 pKa = 3.66EE514 pKa = 4.33FQHH517 pKa = 6.6GLLLTTTKK525 pKa = 10.58FLGLQTIVFEE535 pKa = 4.46RR536 pKa = 11.84NEE538 pKa = 4.44GEE540 pKa = 5.32FILCPHH546 pKa = 7.5LPRR549 pKa = 11.84DD550 pKa = 3.74EE551 pKa = 5.01CVSQLMTPRR560 pKa = 11.84EE561 pKa = 4.33DD562 pKa = 3.3ATGPRR567 pKa = 11.84PSLTASNRR575 pKa = 11.84TLYY578 pKa = 10.85DD579 pKa = 3.14RR580 pKa = 11.84ARR582 pKa = 11.84GLMVTHH588 pKa = 7.42CFTQPDD594 pKa = 3.62VRR596 pKa = 11.84AALYY600 pKa = 10.63NLANQIDD607 pKa = 4.12PTAIALIPQVEE618 pKa = 4.49NGLPPDD624 pKa = 3.89HH625 pKa = 7.08VSLPEE630 pKa = 3.74FSFPDD635 pKa = 3.36SSGFPDD641 pKa = 5.11LSFCLSLYY649 pKa = 9.88STDD652 pKa = 3.48EE653 pKa = 4.12KK654 pKa = 10.6QPPMHH659 pKa = 6.29QFYY662 pKa = 10.65PNISNVHH669 pKa = 6.96AEE671 pKa = 4.02MEE673 pKa = 4.34RR674 pKa = 11.84LRR676 pKa = 11.84EE677 pKa = 3.68AAKK680 pKa = 10.53FKK682 pKa = 10.63FEE684 pKa = 3.87RR685 pKa = 11.84VAVEE689 pKa = 3.86RR690 pKa = 11.84GAIVRR695 pKa = 11.84APSPAPRR702 pKa = 11.84PIDD705 pKa = 4.17PIMEE709 pKa = 4.21VAIGTTGKK717 pKa = 8.42STAVNVPKK725 pKa = 10.74NPPKK729 pKa = 10.15GSKK732 pKa = 8.79TVVVGSDD739 pKa = 3.04ATPPKK744 pKa = 10.33RR745 pKa = 11.84LPTLFMLIRR754 pKa = 11.84KK755 pKa = 8.84IVTRR759 pKa = 11.84RR760 pKa = 11.84GGACTVADD768 pKa = 3.77AAAYY772 pKa = 9.97ASAEE776 pKa = 3.84PRR778 pKa = 11.84RR779 pKa = 11.84VYY781 pKa = 11.0DD782 pKa = 3.38EE783 pKa = 4.41ALNGGLYY790 pKa = 9.28LTGTDD795 pKa = 4.08DD796 pKa = 5.03GDD798 pKa = 4.1IISLNPIATPFPTAATKK815 pKa = 10.15QRR817 pKa = 11.84SDD819 pKa = 3.13EE820 pKa = 4.06SRR822 pKa = 11.84IRR824 pKa = 11.84SLPPRR829 pKa = 11.84ASMAAFNRR837 pKa = 11.84KK838 pKa = 8.5RR839 pKa = 11.84RR840 pKa = 11.84GGPPITFKK848 pKa = 10.8PQLVTINLRR857 pKa = 11.84EE858 pKa = 4.14LYY860 pKa = 10.24GVPLYY865 pKa = 10.49KK866 pKa = 10.92YY867 pKa = 10.63KK868 pKa = 10.68DD869 pKa = 3.85DD870 pKa = 5.94NDD872 pKa = 3.78MLDD875 pKa = 3.15QFTAAIRR882 pKa = 11.84KK883 pKa = 8.45HH884 pKa = 5.6FSHH887 pKa = 6.72LRR889 pKa = 11.84WRR891 pKa = 11.84PILNDD896 pKa = 3.34AKK898 pKa = 10.73HH899 pKa = 5.18LAEE902 pKa = 4.36RR903 pKa = 11.84LEE905 pKa = 3.95VSKK908 pKa = 11.31DD909 pKa = 3.13GLEE912 pKa = 4.37WIDD915 pKa = 4.74AAICEE920 pKa = 4.53CPTVKK925 pKa = 10.61LARR928 pKa = 11.84QSIAWAVYY936 pKa = 8.87LANGYY941 pKa = 9.65EE942 pKa = 4.35IEE944 pKa = 4.48TGPDD948 pKa = 3.13LANKK952 pKa = 7.46TNPDD956 pKa = 3.83PKK958 pKa = 9.92PYY960 pKa = 9.44PALWVPNPNPNPDD973 pKa = 3.99PNGPWEE979 pKa = 4.56DD980 pKa = 3.56WDD982 pKa = 4.15TGPDD986 pKa = 3.67LDD988 pKa = 4.33TQAGYY993 pKa = 10.27AAAEE997 pKa = 4.11PLAAPEE1003 pKa = 4.25IAPLGQPDD1011 pKa = 3.75IPLTSLPRR1019 pKa = 11.84DD1020 pKa = 3.42LQEE1023 pKa = 4.95LSEE1026 pKa = 5.04HH1027 pKa = 6.18YY1028 pKa = 10.69NILQIKK1034 pKa = 9.13LAYY1037 pKa = 10.01LALCNLGVVDD1047 pKa = 4.43PATVILEE1054 pKa = 4.14AVGSSLHH1061 pKa = 5.54PEE1063 pKa = 3.8RR1064 pKa = 11.84RR1065 pKa = 11.84RR1066 pKa = 11.84TARR1069 pKa = 11.84EE1070 pKa = 3.93RR1071 pKa = 11.84IPDD1074 pKa = 3.83TNSAGPAVPKK1084 pKa = 10.35RR1085 pKa = 11.84PPHH1088 pKa = 6.06QKK1090 pKa = 9.4PQSAKK1095 pKa = 9.73PPRR1098 pKa = 11.84KK1099 pKa = 9.02EE1100 pKa = 3.55KK1101 pKa = 10.66TNIEE1105 pKa = 3.99RR1106 pKa = 11.84PLTKK1110 pKa = 9.94NQKK1113 pKa = 8.71IRR1115 pKa = 11.84EE1116 pKa = 4.1RR1117 pKa = 11.84KK1118 pKa = 8.68KK1119 pKa = 10.35RR1120 pKa = 11.84RR1121 pKa = 11.84LAEE1124 pKa = 3.85LRR1126 pKa = 11.84QLEE1129 pKa = 4.32NGAPTKK1135 pKa = 10.83NKK1137 pKa = 10.43SEE1139 pKa = 4.24TSNSSGYY1146 pKa = 11.13ASGDD1150 pKa = 2.94GDD1152 pKa = 4.17GPGKK1156 pKa = 10.56EE1157 pKa = 3.68NVGRR1161 pKa = 11.84HH1162 pKa = 5.28RR1163 pKa = 11.84YY1164 pKa = 4.66WHH1166 pKa = 6.83KK1167 pKa = 9.97EE1168 pKa = 3.87APPAPPPVRR1177 pKa = 11.84NTLRR1181 pKa = 11.84RR1182 pKa = 11.84IWWDD1186 pKa = 3.07SALGDD1191 pKa = 3.84GGLDD1195 pKa = 3.18EE1196 pKa = 4.76TQASRR1201 pKa = 11.84DD1202 pKa = 3.48ADD1204 pKa = 3.6LSHH1207 pKa = 6.71YY1208 pKa = 10.04LQEE1211 pKa = 4.22
MM1 pKa = 7.89DD2 pKa = 5.69ASNPIIPNTLTTMSNILDD20 pKa = 3.53RR21 pKa = 11.84QARR24 pKa = 11.84MTRR27 pKa = 11.84QDD29 pKa = 3.35FSLLRR34 pKa = 11.84DD35 pKa = 3.58RR36 pKa = 11.84LSTQARR42 pKa = 11.84HH43 pKa = 5.57LLEE46 pKa = 4.75VDD48 pKa = 3.83VPRR51 pKa = 11.84LPPEE55 pKa = 3.83QVQEE59 pKa = 3.73ILKK62 pKa = 9.44EE63 pKa = 4.09YY64 pKa = 10.63KK65 pKa = 7.67EE66 pKa = 4.37TRR68 pKa = 11.84VAVPPDD74 pKa = 3.37SKK76 pKa = 11.59APLEE80 pKa = 4.36FPTLDD85 pKa = 4.09LYY87 pKa = 11.47AEE89 pKa = 5.29EE90 pKa = 4.61IPISRR95 pKa = 11.84KK96 pKa = 9.75LDD98 pKa = 3.38FDD100 pKa = 3.6GRR102 pKa = 11.84PIHH105 pKa = 6.66PAGTNSKK112 pKa = 9.75QGRR115 pKa = 11.84VHH117 pKa = 6.85LSKK120 pKa = 11.08SFGSQPRR127 pKa = 11.84PMEE130 pKa = 4.3GVSYY134 pKa = 10.25LAEE137 pKa = 3.77ILGRR141 pKa = 11.84DD142 pKa = 3.7VCNTTHH148 pKa = 7.44IYY150 pKa = 10.29TSGTRR155 pKa = 11.84AGFQKK160 pKa = 10.25RR161 pKa = 11.84VAKK164 pKa = 10.1QLGRR168 pKa = 11.84KK169 pKa = 8.76AVSLAKK175 pKa = 9.69MFRR178 pKa = 11.84EE179 pKa = 4.07VRR181 pKa = 11.84GLGKK185 pKa = 10.23NPTAKK190 pKa = 10.33YY191 pKa = 9.93VNAYY195 pKa = 10.33LDD197 pKa = 3.43TLMPRR202 pKa = 11.84KK203 pKa = 9.67PNTNWPGVHH212 pKa = 6.75TDD214 pKa = 4.63LIDD217 pKa = 5.14GIMDD221 pKa = 5.3GIKK224 pKa = 10.29INFKK228 pKa = 10.55SSAGAPYY235 pKa = 9.91HH236 pKa = 7.12RR237 pKa = 11.84PLHH240 pKa = 6.03LCADD244 pKa = 4.16MVLSTGLPAAIEE256 pKa = 4.24IIKK259 pKa = 10.23NSKK262 pKa = 9.14PIHH265 pKa = 6.73DD266 pKa = 4.1AMEE269 pKa = 5.36DD270 pKa = 3.48DD271 pKa = 4.1VEE273 pKa = 4.46FFISEE278 pKa = 4.58LKK280 pKa = 10.99NKK282 pKa = 7.79TDD284 pKa = 3.35RR285 pKa = 11.84YY286 pKa = 10.56DD287 pKa = 3.42VNEE290 pKa = 4.51LMDD293 pKa = 3.4KK294 pKa = 9.52TRR296 pKa = 11.84PYY298 pKa = 10.81CCLPAHH304 pKa = 6.3WRR306 pKa = 11.84LLFSLVSQGFSATHH320 pKa = 5.26PTFDD324 pKa = 4.51EE325 pKa = 4.25IKK327 pKa = 9.43SANAYY332 pKa = 8.77GHH334 pKa = 6.41SNAHH338 pKa = 5.14GGMRR342 pKa = 11.84RR343 pKa = 11.84RR344 pKa = 11.84YY345 pKa = 8.92DD346 pKa = 2.87WAMTANTRR354 pKa = 11.84GQVIAYY360 pKa = 9.67GDD362 pKa = 3.78DD363 pKa = 3.2ALISIKK369 pKa = 10.35RR370 pKa = 11.84GSDD373 pKa = 2.9IFLVAPDD380 pKa = 4.13FRR382 pKa = 11.84QMDD385 pKa = 4.11GSIHH389 pKa = 6.85RR390 pKa = 11.84EE391 pKa = 3.7DD392 pKa = 2.89IKK394 pKa = 11.54YY395 pKa = 8.86IFRR398 pKa = 11.84WILSHH403 pKa = 7.52LIRR406 pKa = 11.84DD407 pKa = 3.89APGADD412 pKa = 3.33VNFWGPLFSLWEE424 pKa = 4.05FFAIDD429 pKa = 4.92PILMCEE435 pKa = 4.01GSKK438 pKa = 10.72AYY440 pKa = 8.69TKK442 pKa = 10.54KK443 pKa = 10.8NPDD446 pKa = 3.17AGMLSGVPGTTLINTVKK463 pKa = 10.7CATAWDD469 pKa = 4.92LYY471 pKa = 11.19LEE473 pKa = 4.69HH474 pKa = 7.19CANTGSDD481 pKa = 3.74ALDD484 pKa = 3.98PDD486 pKa = 3.52MARR489 pKa = 11.84RR490 pKa = 11.84FMAKK494 pKa = 9.75HH495 pKa = 5.24GLEE498 pKa = 4.2LKK500 pKa = 10.34PGTFSPQRR508 pKa = 11.84LPTPDD513 pKa = 3.66EE514 pKa = 4.33FQHH517 pKa = 6.6GLLLTTTKK525 pKa = 10.58FLGLQTIVFEE535 pKa = 4.46RR536 pKa = 11.84NEE538 pKa = 4.44GEE540 pKa = 5.32FILCPHH546 pKa = 7.5LPRR549 pKa = 11.84DD550 pKa = 3.74EE551 pKa = 5.01CVSQLMTPRR560 pKa = 11.84EE561 pKa = 4.33DD562 pKa = 3.3ATGPRR567 pKa = 11.84PSLTASNRR575 pKa = 11.84TLYY578 pKa = 10.85DD579 pKa = 3.14RR580 pKa = 11.84ARR582 pKa = 11.84GLMVTHH588 pKa = 7.42CFTQPDD594 pKa = 3.62VRR596 pKa = 11.84AALYY600 pKa = 10.63NLANQIDD607 pKa = 4.12PTAIALIPQVEE618 pKa = 4.49NGLPPDD624 pKa = 3.89HH625 pKa = 7.08VSLPEE630 pKa = 3.74FSFPDD635 pKa = 3.36SSGFPDD641 pKa = 5.11LSFCLSLYY649 pKa = 9.88STDD652 pKa = 3.48EE653 pKa = 4.12KK654 pKa = 10.6QPPMHH659 pKa = 6.29QFYY662 pKa = 10.65PNISNVHH669 pKa = 6.96AEE671 pKa = 4.02MEE673 pKa = 4.34RR674 pKa = 11.84LRR676 pKa = 11.84EE677 pKa = 3.68AAKK680 pKa = 10.53FKK682 pKa = 10.63FEE684 pKa = 3.87RR685 pKa = 11.84VAVEE689 pKa = 3.86RR690 pKa = 11.84GAIVRR695 pKa = 11.84APSPAPRR702 pKa = 11.84PIDD705 pKa = 4.17PIMEE709 pKa = 4.21VAIGTTGKK717 pKa = 8.42STAVNVPKK725 pKa = 10.74NPPKK729 pKa = 10.15GSKK732 pKa = 8.79TVVVGSDD739 pKa = 3.04ATPPKK744 pKa = 10.33RR745 pKa = 11.84LPTLFMLIRR754 pKa = 11.84KK755 pKa = 8.84IVTRR759 pKa = 11.84RR760 pKa = 11.84GGACTVADD768 pKa = 3.77AAAYY772 pKa = 9.97ASAEE776 pKa = 3.84PRR778 pKa = 11.84RR779 pKa = 11.84VYY781 pKa = 11.0DD782 pKa = 3.38EE783 pKa = 4.41ALNGGLYY790 pKa = 9.28LTGTDD795 pKa = 4.08DD796 pKa = 5.03GDD798 pKa = 4.1IISLNPIATPFPTAATKK815 pKa = 10.15QRR817 pKa = 11.84SDD819 pKa = 3.13EE820 pKa = 4.06SRR822 pKa = 11.84IRR824 pKa = 11.84SLPPRR829 pKa = 11.84ASMAAFNRR837 pKa = 11.84KK838 pKa = 8.5RR839 pKa = 11.84RR840 pKa = 11.84GGPPITFKK848 pKa = 10.8PQLVTINLRR857 pKa = 11.84EE858 pKa = 4.14LYY860 pKa = 10.24GVPLYY865 pKa = 10.49KK866 pKa = 10.92YY867 pKa = 10.63KK868 pKa = 10.68DD869 pKa = 3.85DD870 pKa = 5.94NDD872 pKa = 3.78MLDD875 pKa = 3.15QFTAAIRR882 pKa = 11.84KK883 pKa = 8.45HH884 pKa = 5.6FSHH887 pKa = 6.72LRR889 pKa = 11.84WRR891 pKa = 11.84PILNDD896 pKa = 3.34AKK898 pKa = 10.73HH899 pKa = 5.18LAEE902 pKa = 4.36RR903 pKa = 11.84LEE905 pKa = 3.95VSKK908 pKa = 11.31DD909 pKa = 3.13GLEE912 pKa = 4.37WIDD915 pKa = 4.74AAICEE920 pKa = 4.53CPTVKK925 pKa = 10.61LARR928 pKa = 11.84QSIAWAVYY936 pKa = 8.87LANGYY941 pKa = 9.65EE942 pKa = 4.35IEE944 pKa = 4.48TGPDD948 pKa = 3.13LANKK952 pKa = 7.46TNPDD956 pKa = 3.83PKK958 pKa = 9.92PYY960 pKa = 9.44PALWVPNPNPNPDD973 pKa = 3.99PNGPWEE979 pKa = 4.56DD980 pKa = 3.56WDD982 pKa = 4.15TGPDD986 pKa = 3.67LDD988 pKa = 4.33TQAGYY993 pKa = 10.27AAAEE997 pKa = 4.11PLAAPEE1003 pKa = 4.25IAPLGQPDD1011 pKa = 3.75IPLTSLPRR1019 pKa = 11.84DD1020 pKa = 3.42LQEE1023 pKa = 4.95LSEE1026 pKa = 5.04HH1027 pKa = 6.18YY1028 pKa = 10.69NILQIKK1034 pKa = 9.13LAYY1037 pKa = 10.01LALCNLGVVDD1047 pKa = 4.43PATVILEE1054 pKa = 4.14AVGSSLHH1061 pKa = 5.54PEE1063 pKa = 3.8RR1064 pKa = 11.84RR1065 pKa = 11.84RR1066 pKa = 11.84TARR1069 pKa = 11.84EE1070 pKa = 3.93RR1071 pKa = 11.84IPDD1074 pKa = 3.83TNSAGPAVPKK1084 pKa = 10.35RR1085 pKa = 11.84PPHH1088 pKa = 6.06QKK1090 pKa = 9.4PQSAKK1095 pKa = 9.73PPRR1098 pKa = 11.84KK1099 pKa = 9.02EE1100 pKa = 3.55KK1101 pKa = 10.66TNIEE1105 pKa = 3.99RR1106 pKa = 11.84PLTKK1110 pKa = 9.94NQKK1113 pKa = 8.71IRR1115 pKa = 11.84EE1116 pKa = 4.1RR1117 pKa = 11.84KK1118 pKa = 8.68KK1119 pKa = 10.35RR1120 pKa = 11.84RR1121 pKa = 11.84LAEE1124 pKa = 3.85LRR1126 pKa = 11.84QLEE1129 pKa = 4.32NGAPTKK1135 pKa = 10.83NKK1137 pKa = 10.43SEE1139 pKa = 4.24TSNSSGYY1146 pKa = 11.13ASGDD1150 pKa = 2.94GDD1152 pKa = 4.17GPGKK1156 pKa = 10.56EE1157 pKa = 3.68NVGRR1161 pKa = 11.84HH1162 pKa = 5.28RR1163 pKa = 11.84YY1164 pKa = 4.66WHH1166 pKa = 6.83KK1167 pKa = 9.97EE1168 pKa = 3.87APPAPPPVRR1177 pKa = 11.84NTLRR1181 pKa = 11.84RR1182 pKa = 11.84IWWDD1186 pKa = 3.07SALGDD1191 pKa = 3.84GGLDD1195 pKa = 3.18EE1196 pKa = 4.76TQASRR1201 pKa = 11.84DD1202 pKa = 3.48ADD1204 pKa = 3.6LSHH1207 pKa = 6.71YY1208 pKa = 10.04LQEE1211 pKa = 4.22
Molecular weight: 134.85 kDa
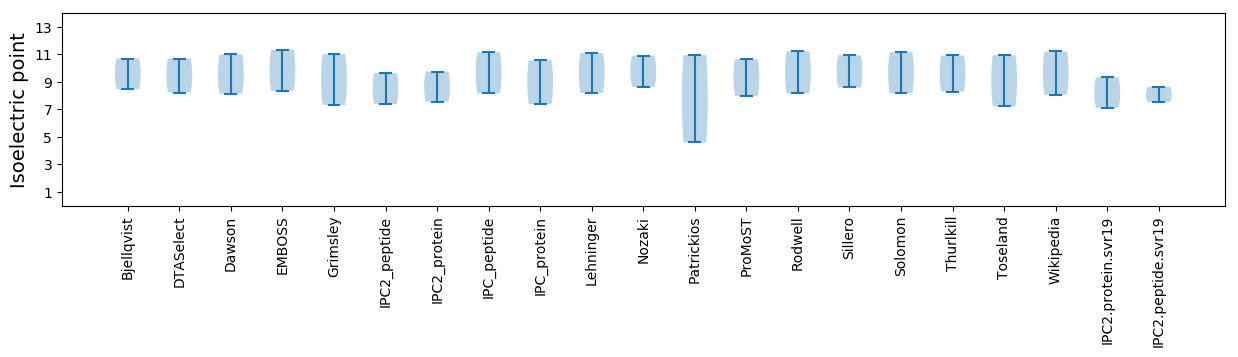
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHW9|A0A1L3KHW9_9VIRU RdRp OS=Hubei permutotetra-like virus 7 OX=1923081 PE=4 SV=1
MM1 pKa = 6.7VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84AKK10 pKa = 9.62PATPAAMQAATVMALAKK27 pKa = 10.31KK28 pKa = 8.96MSAVTVTGTKK38 pKa = 8.98RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84SGIPSGVSGGIQRR59 pKa = 11.84SAMEE63 pKa = 3.97GSTRR67 pKa = 11.84LKK69 pKa = 9.22RR70 pKa = 11.84HH71 pKa = 5.58EE72 pKa = 4.54MLTSLTISKK81 pKa = 10.67NSTHH85 pKa = 6.02LVKK88 pKa = 10.16TIAIQPSSFSLLKK101 pKa = 10.92GIAKK105 pKa = 10.12SFEE108 pKa = 3.86RR109 pKa = 11.84TVWHH113 pKa = 6.64SLTIFYY119 pKa = 10.23KK120 pKa = 10.39PAVATTFGGLIAYY133 pKa = 8.97GIDD136 pKa = 3.03WSSKK140 pKa = 9.32VGTDD144 pKa = 3.09ADD146 pKa = 4.18RR147 pKa = 11.84TKK149 pKa = 10.74VCSLTPVLTHH159 pKa = 6.65AAWMDD164 pKa = 3.7SEE166 pKa = 4.35RR167 pKa = 11.84APLRR171 pKa = 11.84VPKK174 pKa = 10.44KK175 pKa = 9.6LLQSRR180 pKa = 11.84RR181 pKa = 11.84FYY183 pKa = 11.05INGADD188 pKa = 4.47ADD190 pKa = 4.18DD191 pKa = 4.98AGPGNIVVSIDD202 pKa = 3.14ITASSADD209 pKa = 3.63VVVGEE214 pKa = 4.85LWCQYY219 pKa = 9.98DD220 pKa = 3.6VEE222 pKa = 4.6MTGTVVPP229 pKa = 4.81
MM1 pKa = 6.7VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84AKK10 pKa = 9.62PATPAAMQAATVMALAKK27 pKa = 10.31KK28 pKa = 8.96MSAVTVTGTKK38 pKa = 8.98RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84SGIPSGVSGGIQRR59 pKa = 11.84SAMEE63 pKa = 3.97GSTRR67 pKa = 11.84LKK69 pKa = 9.22RR70 pKa = 11.84HH71 pKa = 5.58EE72 pKa = 4.54MLTSLTISKK81 pKa = 10.67NSTHH85 pKa = 6.02LVKK88 pKa = 10.16TIAIQPSSFSLLKK101 pKa = 10.92GIAKK105 pKa = 10.12SFEE108 pKa = 3.86RR109 pKa = 11.84TVWHH113 pKa = 6.64SLTIFYY119 pKa = 10.23KK120 pKa = 10.39PAVATTFGGLIAYY133 pKa = 8.97GIDD136 pKa = 3.03WSSKK140 pKa = 9.32VGTDD144 pKa = 3.09ADD146 pKa = 4.18RR147 pKa = 11.84TKK149 pKa = 10.74VCSLTPVLTHH159 pKa = 6.65AAWMDD164 pKa = 3.7SEE166 pKa = 4.35RR167 pKa = 11.84APLRR171 pKa = 11.84VPKK174 pKa = 10.44KK175 pKa = 9.6LLQSRR180 pKa = 11.84RR181 pKa = 11.84FYY183 pKa = 11.05INGADD188 pKa = 4.47ADD190 pKa = 4.18DD191 pKa = 4.98AGPGNIVVSIDD202 pKa = 3.14ITASSADD209 pKa = 3.63VVVGEE214 pKa = 4.85LWCQYY219 pKa = 9.98DD220 pKa = 3.6VEE222 pKa = 4.6MTGTVVPP229 pKa = 4.81
Molecular weight: 24.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1440 |
229 |
1211 |
720.0 |
79.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.167 ± 0.644 | 1.181 ± 0.151 |
6.25 ± 0.923 | 4.931 ± 1.133 |
3.056 ± 0.428 | 6.25 ± 0.361 |
2.431 ± 0.335 | 5.347 ± 0.162 |
5.417 ± 0.342 | 8.819 ± 1.112 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.292 ± 0.589 | 3.819 ± 1.23 |
8.472 ± 1.798 | 2.778 ± 0.291 |
7.569 ± 0.785 | 6.389 ± 1.577 |
6.667 ± 1.441 | 5.139 ± 1.762 |
1.389 ± 0.175 | 2.639 ± 0.437 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
