
Sewage-associated circular DNA virus-28
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.82
Get precalculated fractions of proteins
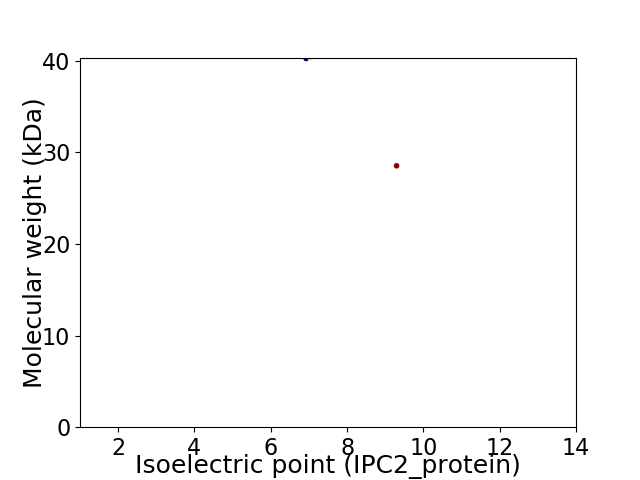
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI50|A0A0B4UI50_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-28 OX=1592095 PE=3 SV=1
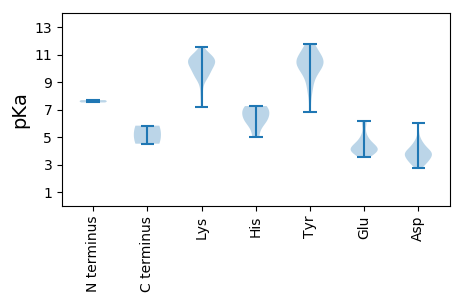
MM1 pKa = 7.51EE2 pKa = 5.94APPPAYY8 pKa = 10.11SPPPPSVTPPIEE20 pKa = 3.79SSRR23 pKa = 11.84PFRR26 pKa = 11.84LAGKK30 pKa = 10.05AIFLTWPQNDD40 pKa = 3.12ITKK43 pKa = 9.98EE44 pKa = 3.72DD45 pKa = 3.74LMAKK49 pKa = 10.15LVSLWEE55 pKa = 4.59AKK57 pKa = 10.55LSWAVVAEE65 pKa = 4.39EE66 pKa = 4.18SHH68 pKa = 7.01KK69 pKa = 11.02SGEE72 pKa = 4.21PHH74 pKa = 5.03VHH76 pKa = 7.26AIAQFTEE83 pKa = 5.05RR84 pKa = 11.84IDD86 pKa = 3.89LKK88 pKa = 10.96NANPVLDD95 pKa = 4.42ALTGKK100 pKa = 9.52HH101 pKa = 6.22GNYY104 pKa = 9.84QSVKK108 pKa = 9.21SAKK111 pKa = 9.23KK112 pKa = 9.2VLRR115 pKa = 11.84YY116 pKa = 9.18VCKK119 pKa = 9.43DD120 pKa = 3.09GQYY123 pKa = 8.29ITHH126 pKa = 6.75GEE128 pKa = 4.15VPDD131 pKa = 4.09FAEE134 pKa = 4.7KK135 pKa = 10.82AKK137 pKa = 10.76LQDD140 pKa = 3.17WAAKK144 pKa = 10.44LIMEE148 pKa = 4.62EE149 pKa = 3.84QATYY153 pKa = 10.76RR154 pKa = 11.84DD155 pKa = 4.06LVRR158 pKa = 11.84QQPGYY163 pKa = 11.6CMLQKK168 pKa = 10.53PKK170 pKa = 10.8LEE172 pKa = 4.13EE173 pKa = 4.15FIGWSKK179 pKa = 10.46RR180 pKa = 11.84QRR182 pKa = 11.84LADD185 pKa = 3.77SLLPWRR191 pKa = 11.84VLTVKK196 pKa = 10.46PNAPYY201 pKa = 10.37VHH203 pKa = 6.02QALWTFLRR211 pKa = 11.84EE212 pKa = 3.99NVLVPRR218 pKa = 11.84TPRR221 pKa = 11.84QAQLWLSGLPGVGKK235 pKa = 8.43TRR237 pKa = 11.84FLSYY241 pKa = 10.42LRR243 pKa = 11.84ARR245 pKa = 11.84LRR247 pKa = 11.84VYY249 pKa = 11.14DD250 pKa = 3.82MPRR253 pKa = 11.84DD254 pKa = 3.33EE255 pKa = 6.07DD256 pKa = 4.6FYY258 pKa = 11.64DD259 pKa = 4.55DD260 pKa = 6.03FEE262 pKa = 6.19DD263 pKa = 4.19GCFDD267 pKa = 5.14LVVLDD272 pKa = 4.06EE273 pKa = 5.2FKK275 pKa = 10.97AHH277 pKa = 7.15KK278 pKa = 10.11KK279 pKa = 9.6IQFLNAWADD288 pKa = 3.87GQPLPLRR295 pKa = 11.84KK296 pKa = 9.62KK297 pKa = 10.06GSQSVKK303 pKa = 9.84TDD305 pKa = 3.59NLPLIIVSNYY315 pKa = 9.43TIEE318 pKa = 4.05EE319 pKa = 4.69CYY321 pKa = 10.41KK322 pKa = 10.85SGVGRR327 pKa = 11.84DD328 pKa = 3.46ALVDD332 pKa = 3.44RR333 pKa = 11.84FTQVFVDD340 pKa = 3.93SLFSIDD346 pKa = 4.32DD347 pKa = 3.93PLYY350 pKa = 10.48TEE352 pKa = 4.51
MM1 pKa = 7.51EE2 pKa = 5.94APPPAYY8 pKa = 10.11SPPPPSVTPPIEE20 pKa = 3.79SSRR23 pKa = 11.84PFRR26 pKa = 11.84LAGKK30 pKa = 10.05AIFLTWPQNDD40 pKa = 3.12ITKK43 pKa = 9.98EE44 pKa = 3.72DD45 pKa = 3.74LMAKK49 pKa = 10.15LVSLWEE55 pKa = 4.59AKK57 pKa = 10.55LSWAVVAEE65 pKa = 4.39EE66 pKa = 4.18SHH68 pKa = 7.01KK69 pKa = 11.02SGEE72 pKa = 4.21PHH74 pKa = 5.03VHH76 pKa = 7.26AIAQFTEE83 pKa = 5.05RR84 pKa = 11.84IDD86 pKa = 3.89LKK88 pKa = 10.96NANPVLDD95 pKa = 4.42ALTGKK100 pKa = 9.52HH101 pKa = 6.22GNYY104 pKa = 9.84QSVKK108 pKa = 9.21SAKK111 pKa = 9.23KK112 pKa = 9.2VLRR115 pKa = 11.84YY116 pKa = 9.18VCKK119 pKa = 9.43DD120 pKa = 3.09GQYY123 pKa = 8.29ITHH126 pKa = 6.75GEE128 pKa = 4.15VPDD131 pKa = 4.09FAEE134 pKa = 4.7KK135 pKa = 10.82AKK137 pKa = 10.76LQDD140 pKa = 3.17WAAKK144 pKa = 10.44LIMEE148 pKa = 4.62EE149 pKa = 3.84QATYY153 pKa = 10.76RR154 pKa = 11.84DD155 pKa = 4.06LVRR158 pKa = 11.84QQPGYY163 pKa = 11.6CMLQKK168 pKa = 10.53PKK170 pKa = 10.8LEE172 pKa = 4.13EE173 pKa = 4.15FIGWSKK179 pKa = 10.46RR180 pKa = 11.84QRR182 pKa = 11.84LADD185 pKa = 3.77SLLPWRR191 pKa = 11.84VLTVKK196 pKa = 10.46PNAPYY201 pKa = 10.37VHH203 pKa = 6.02QALWTFLRR211 pKa = 11.84EE212 pKa = 3.99NVLVPRR218 pKa = 11.84TPRR221 pKa = 11.84QAQLWLSGLPGVGKK235 pKa = 8.43TRR237 pKa = 11.84FLSYY241 pKa = 10.42LRR243 pKa = 11.84ARR245 pKa = 11.84LRR247 pKa = 11.84VYY249 pKa = 11.14DD250 pKa = 3.82MPRR253 pKa = 11.84DD254 pKa = 3.33EE255 pKa = 6.07DD256 pKa = 4.6FYY258 pKa = 11.64DD259 pKa = 4.55DD260 pKa = 6.03FEE262 pKa = 6.19DD263 pKa = 4.19GCFDD267 pKa = 5.14LVVLDD272 pKa = 4.06EE273 pKa = 5.2FKK275 pKa = 10.97AHH277 pKa = 7.15KK278 pKa = 10.11KK279 pKa = 9.6IQFLNAWADD288 pKa = 3.87GQPLPLRR295 pKa = 11.84KK296 pKa = 9.62KK297 pKa = 10.06GSQSVKK303 pKa = 9.84TDD305 pKa = 3.59NLPLIIVSNYY315 pKa = 9.43TIEE318 pKa = 4.05EE319 pKa = 4.69CYY321 pKa = 10.41KK322 pKa = 10.85SGVGRR327 pKa = 11.84DD328 pKa = 3.46ALVDD332 pKa = 3.44RR333 pKa = 11.84FTQVFVDD340 pKa = 3.93SLFSIDD346 pKa = 4.32DD347 pKa = 3.93PLYY350 pKa = 10.48TEE352 pKa = 4.51
Molecular weight: 40.25 kDa
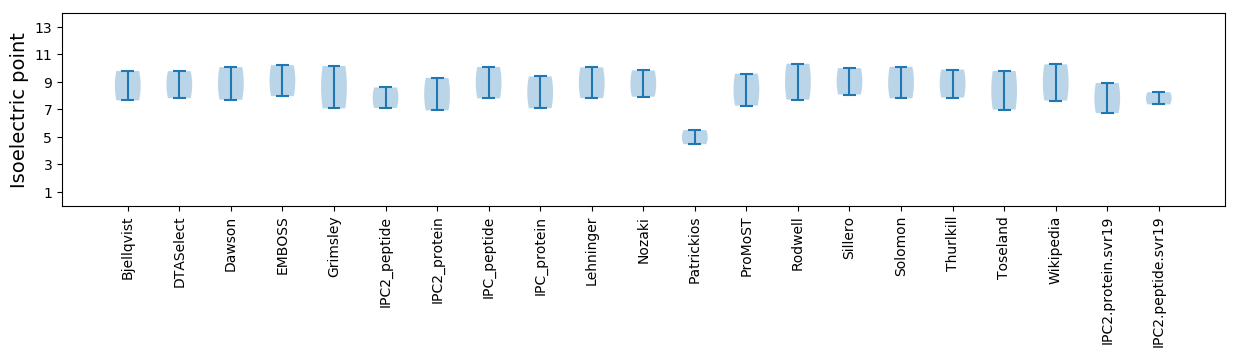
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI50|A0A0B4UI50_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-28 OX=1592095 PE=3 SV=1
MM1 pKa = 7.68SYY3 pKa = 10.45LGKK6 pKa = 10.43RR7 pKa = 11.84KK8 pKa = 7.22MTSYY12 pKa = 8.5PTARR16 pKa = 11.84KK17 pKa = 9.54VSRR20 pKa = 11.84QRR22 pKa = 11.84SPLTLQYY29 pKa = 10.2GVPVTIPAGARR40 pKa = 11.84GYY42 pKa = 10.29VRR44 pKa = 11.84NQGRR48 pKa = 11.84FGRR51 pKa = 11.84TARR54 pKa = 11.84MQRR57 pKa = 11.84GDD59 pKa = 3.56SMPEE63 pKa = 3.51MKK65 pKa = 10.79YY66 pKa = 10.92LDD68 pKa = 3.78TSLTWSCDD76 pKa = 3.08TTGEE80 pKa = 4.11VSAPYY85 pKa = 10.73LNIIPQGVGEE95 pKa = 4.1NQRR98 pKa = 11.84VGRR101 pKa = 11.84KK102 pKa = 7.99VVVRR106 pKa = 11.84SIGIKK111 pKa = 10.45GLVVPLNTSVDD122 pKa = 3.17ASLIKK127 pKa = 10.36IQVVLDD133 pKa = 3.67TQANGTAPTFGGSADD148 pKa = 4.62SIWEE152 pKa = 4.31SNSIHH157 pKa = 6.33SWRR160 pKa = 11.84NPDD163 pKa = 2.73NAARR167 pKa = 11.84FKK169 pKa = 10.81VIKK172 pKa = 10.39EE173 pKa = 3.9FDD175 pKa = 3.7LKK177 pKa = 10.33PTVTAAYY184 pKa = 9.64YY185 pKa = 10.88NSTTTTEE192 pKa = 3.87KK193 pKa = 10.78RR194 pKa = 11.84LTNILRR200 pKa = 11.84VEE202 pKa = 4.0WFKK205 pKa = 11.55KK206 pKa = 9.79CFIPIEE212 pKa = 4.17YY213 pKa = 10.33DD214 pKa = 3.44SVLTTGALTTVRR226 pKa = 11.84SNNLLITCINSANDD240 pKa = 3.27DD241 pKa = 3.78QYY243 pKa = 11.78NFTGMQRR250 pKa = 11.84IRR252 pKa = 11.84YY253 pKa = 6.81TDD255 pKa = 2.88AA256 pKa = 5.83
MM1 pKa = 7.68SYY3 pKa = 10.45LGKK6 pKa = 10.43RR7 pKa = 11.84KK8 pKa = 7.22MTSYY12 pKa = 8.5PTARR16 pKa = 11.84KK17 pKa = 9.54VSRR20 pKa = 11.84QRR22 pKa = 11.84SPLTLQYY29 pKa = 10.2GVPVTIPAGARR40 pKa = 11.84GYY42 pKa = 10.29VRR44 pKa = 11.84NQGRR48 pKa = 11.84FGRR51 pKa = 11.84TARR54 pKa = 11.84MQRR57 pKa = 11.84GDD59 pKa = 3.56SMPEE63 pKa = 3.51MKK65 pKa = 10.79YY66 pKa = 10.92LDD68 pKa = 3.78TSLTWSCDD76 pKa = 3.08TTGEE80 pKa = 4.11VSAPYY85 pKa = 10.73LNIIPQGVGEE95 pKa = 4.1NQRR98 pKa = 11.84VGRR101 pKa = 11.84KK102 pKa = 7.99VVVRR106 pKa = 11.84SIGIKK111 pKa = 10.45GLVVPLNTSVDD122 pKa = 3.17ASLIKK127 pKa = 10.36IQVVLDD133 pKa = 3.67TQANGTAPTFGGSADD148 pKa = 4.62SIWEE152 pKa = 4.31SNSIHH157 pKa = 6.33SWRR160 pKa = 11.84NPDD163 pKa = 2.73NAARR167 pKa = 11.84FKK169 pKa = 10.81VIKK172 pKa = 10.39EE173 pKa = 3.9FDD175 pKa = 3.7LKK177 pKa = 10.33PTVTAAYY184 pKa = 9.64YY185 pKa = 10.88NSTTTTEE192 pKa = 3.87KK193 pKa = 10.78RR194 pKa = 11.84LTNILRR200 pKa = 11.84VEE202 pKa = 4.0WFKK205 pKa = 11.55KK206 pKa = 9.79CFIPIEE212 pKa = 4.17YY213 pKa = 10.33DD214 pKa = 3.44SVLTTGALTTVRR226 pKa = 11.84SNNLLITCINSANDD240 pKa = 3.27DD241 pKa = 3.78QYY243 pKa = 11.78NFTGMQRR250 pKa = 11.84IRR252 pKa = 11.84YY253 pKa = 6.81TDD255 pKa = 2.88AA256 pKa = 5.83
Molecular weight: 28.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608 |
256 |
352 |
304.0 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.072 ± 0.474 | 1.151 ± 0.012 |
5.921 ± 0.712 | 4.77 ± 0.949 |
3.618 ± 0.51 | 5.592 ± 0.83 |
1.316 ± 0.534 | 4.77 ± 0.854 |
6.414 ± 0.771 | 9.046 ± 1.388 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.809 ± 0.308 | 3.947 ± 1.103 |
6.414 ± 0.996 | 4.441 ± 0.308 |
6.579 ± 0.712 | 6.579 ± 0.712 |
6.908 ± 2.099 | 7.566 ± 0.142 |
2.138 ± 0.332 | 3.947 ± 0.202 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
