
Tuber aestivum virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Totivirus
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
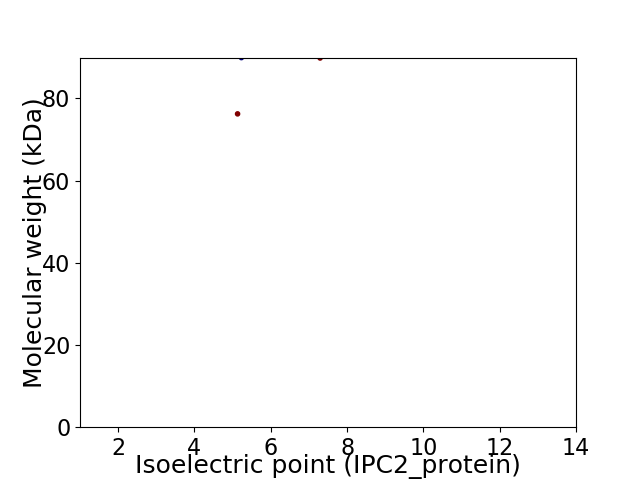
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E5KIN7|E5KIN7_9VIRU RNA-directed RNA polymerase OS=Tuber aestivum virus 1 OX=927810 PE=3 SV=1
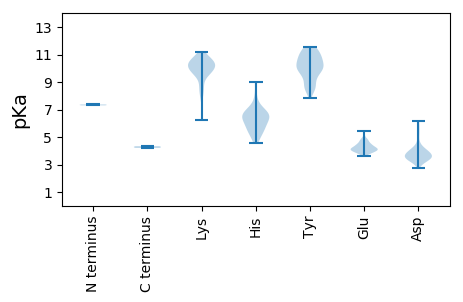
MM1 pKa = 7.3YY2 pKa = 8.49KK3 pKa = 9.5TFNSLSEE10 pKa = 4.04NAPVSTFEE18 pKa = 4.51TIQSRR23 pKa = 11.84GTTSLMTRR31 pKa = 11.84LRR33 pKa = 11.84LNLAYY38 pKa = 10.56DD39 pKa = 3.55NLDD42 pKa = 3.41FNRR45 pKa = 11.84VYY47 pKa = 10.77SSKK50 pKa = 10.62QDD52 pKa = 3.58YY53 pKa = 8.42TWLGKK58 pKa = 9.73AAAFIVEE65 pKa = 4.79AEE67 pKa = 4.26LSLDD71 pKa = 4.64GIAKK75 pKa = 10.04QYY77 pKa = 8.86LTLDD81 pKa = 3.36GAINFDD87 pKa = 3.49NVLAEE92 pKa = 4.14LRR94 pKa = 11.84TGAGLQTTNVSNHH107 pKa = 5.19SVSVSSWRR115 pKa = 11.84WYY117 pKa = 10.13DD118 pKa = 3.23NHH120 pKa = 5.66VTLLFNLLRR129 pKa = 11.84LYY131 pKa = 11.41VMADD135 pKa = 3.61LQLKK139 pKa = 10.29GPITTGSYY147 pKa = 9.95PKK149 pKa = 10.47YY150 pKa = 10.29DD151 pKa = 4.33DD152 pKa = 3.4GHH154 pKa = 7.98VEE156 pKa = 3.63IDD158 pKa = 3.99LNAGLPLPDD167 pKa = 3.93EE168 pKa = 4.79TIVWSWPGSKK178 pKa = 10.43NSLNYY183 pKa = 9.69PEE185 pKa = 4.64WGNFTEE191 pKa = 4.88YY192 pKa = 10.38MPLNDD197 pKa = 3.72EE198 pKa = 4.75AYY200 pKa = 9.54IDD202 pKa = 3.84VGGLLPEE209 pKa = 4.08EE210 pKa = 4.29VKK212 pKa = 10.18MVLMATGAWEE222 pKa = 3.8RR223 pKa = 11.84QTNFKK228 pKa = 10.9LDD230 pKa = 3.57FPTPKK235 pKa = 9.88LAEE238 pKa = 4.55RR239 pKa = 11.84IYY241 pKa = 10.78YY242 pKa = 9.83RR243 pKa = 11.84YY244 pKa = 8.63PSKK247 pKa = 10.6VTMIDD252 pKa = 3.04DD253 pKa = 3.81WLDD256 pKa = 3.56GEE258 pKa = 4.77IPDD261 pKa = 5.38INFGLPNPKK270 pKa = 9.78VVWNAIRR277 pKa = 11.84KK278 pKa = 8.49YY279 pKa = 9.01VTTNNLYY286 pKa = 9.75NQFYY290 pKa = 10.21SAAAIINQVMLTPLPDD306 pKa = 3.55SAEE309 pKa = 3.91GMSWLIHH316 pKa = 5.33EE317 pKa = 4.83AQVVLPKK324 pKa = 10.42FGSVRR329 pKa = 11.84GRR331 pKa = 11.84YY332 pKa = 8.49PFLLRR337 pKa = 11.84GEE339 pKa = 4.45GALIQAKK346 pKa = 10.16ALEE349 pKa = 4.83DD350 pKa = 3.14WTMLQTNPNLLFSTSMVTASLFNIGLAVRR379 pKa = 11.84DD380 pKa = 3.67SMLRR384 pKa = 11.84GIANDD389 pKa = 3.58SLSGVDD395 pKa = 3.37TSLFQQPEE403 pKa = 4.44LAWAAAVSLACGIDD417 pKa = 3.47VPLNGMSNCYY427 pKa = 10.04VYY429 pKa = 11.15YY430 pKa = 8.46PTLADD435 pKa = 3.68SNVRR439 pKa = 11.84LALPALIKK447 pKa = 9.86TPRR450 pKa = 11.84GYY452 pKa = 9.89WIEE455 pKa = 4.11GDD457 pKa = 3.96SIVIDD462 pKa = 3.55GVPFVGSPYY471 pKa = 10.62VLYY474 pKa = 10.02PLAVYY479 pKa = 10.45DD480 pKa = 4.11FANPYY485 pKa = 10.02SGAFALPKK493 pKa = 9.79PIRR496 pKa = 11.84RR497 pKa = 11.84TRR499 pKa = 11.84KK500 pKa = 7.97GAVFTVIGAWMYY512 pKa = 10.84SWVARR517 pKa = 11.84LAGYY521 pKa = 9.44DD522 pKa = 3.15VHH524 pKa = 9.03ISVNGSNVDD533 pKa = 3.42YY534 pKa = 11.1YY535 pKa = 11.46KK536 pKa = 11.17YY537 pKa = 10.31FASNEE542 pKa = 4.06NSWVHH547 pKa = 5.6PLWNGISDD555 pKa = 4.03EE556 pKa = 4.24IEE558 pKa = 4.11GVFVHH563 pKa = 7.25AIVRR567 pKa = 11.84RR568 pKa = 11.84NKK570 pKa = 10.2HH571 pKa = 5.78FLDD574 pKa = 3.85IPDD577 pKa = 3.43YY578 pKa = 9.86TIPGNLTDD586 pKa = 3.43VSVSVKK592 pKa = 10.33ILDD595 pKa = 3.95RR596 pKa = 11.84YY597 pKa = 10.69VSDD600 pKa = 4.16SSLTTKK606 pKa = 10.21FRR608 pKa = 11.84ANYY611 pKa = 9.2NVGSVTVPKK620 pKa = 10.61SGIQVVTSGDD630 pKa = 3.31LRR632 pKa = 11.84KK633 pKa = 9.37FWGYY637 pKa = 7.85VRR639 pKa = 11.84RR640 pKa = 11.84SQPGLTMVGMAQPAIMPQPKK660 pKa = 9.05HH661 pKa = 5.55QLVEE665 pKa = 3.95IDD667 pKa = 3.88QEE669 pKa = 4.42PEE671 pKa = 3.84VTSEE675 pKa = 3.92MVNMIEE681 pKa = 4.33
MM1 pKa = 7.3YY2 pKa = 8.49KK3 pKa = 9.5TFNSLSEE10 pKa = 4.04NAPVSTFEE18 pKa = 4.51TIQSRR23 pKa = 11.84GTTSLMTRR31 pKa = 11.84LRR33 pKa = 11.84LNLAYY38 pKa = 10.56DD39 pKa = 3.55NLDD42 pKa = 3.41FNRR45 pKa = 11.84VYY47 pKa = 10.77SSKK50 pKa = 10.62QDD52 pKa = 3.58YY53 pKa = 8.42TWLGKK58 pKa = 9.73AAAFIVEE65 pKa = 4.79AEE67 pKa = 4.26LSLDD71 pKa = 4.64GIAKK75 pKa = 10.04QYY77 pKa = 8.86LTLDD81 pKa = 3.36GAINFDD87 pKa = 3.49NVLAEE92 pKa = 4.14LRR94 pKa = 11.84TGAGLQTTNVSNHH107 pKa = 5.19SVSVSSWRR115 pKa = 11.84WYY117 pKa = 10.13DD118 pKa = 3.23NHH120 pKa = 5.66VTLLFNLLRR129 pKa = 11.84LYY131 pKa = 11.41VMADD135 pKa = 3.61LQLKK139 pKa = 10.29GPITTGSYY147 pKa = 9.95PKK149 pKa = 10.47YY150 pKa = 10.29DD151 pKa = 4.33DD152 pKa = 3.4GHH154 pKa = 7.98VEE156 pKa = 3.63IDD158 pKa = 3.99LNAGLPLPDD167 pKa = 3.93EE168 pKa = 4.79TIVWSWPGSKK178 pKa = 10.43NSLNYY183 pKa = 9.69PEE185 pKa = 4.64WGNFTEE191 pKa = 4.88YY192 pKa = 10.38MPLNDD197 pKa = 3.72EE198 pKa = 4.75AYY200 pKa = 9.54IDD202 pKa = 3.84VGGLLPEE209 pKa = 4.08EE210 pKa = 4.29VKK212 pKa = 10.18MVLMATGAWEE222 pKa = 3.8RR223 pKa = 11.84QTNFKK228 pKa = 10.9LDD230 pKa = 3.57FPTPKK235 pKa = 9.88LAEE238 pKa = 4.55RR239 pKa = 11.84IYY241 pKa = 10.78YY242 pKa = 9.83RR243 pKa = 11.84YY244 pKa = 8.63PSKK247 pKa = 10.6VTMIDD252 pKa = 3.04DD253 pKa = 3.81WLDD256 pKa = 3.56GEE258 pKa = 4.77IPDD261 pKa = 5.38INFGLPNPKK270 pKa = 9.78VVWNAIRR277 pKa = 11.84KK278 pKa = 8.49YY279 pKa = 9.01VTTNNLYY286 pKa = 9.75NQFYY290 pKa = 10.21SAAAIINQVMLTPLPDD306 pKa = 3.55SAEE309 pKa = 3.91GMSWLIHH316 pKa = 5.33EE317 pKa = 4.83AQVVLPKK324 pKa = 10.42FGSVRR329 pKa = 11.84GRR331 pKa = 11.84YY332 pKa = 8.49PFLLRR337 pKa = 11.84GEE339 pKa = 4.45GALIQAKK346 pKa = 10.16ALEE349 pKa = 4.83DD350 pKa = 3.14WTMLQTNPNLLFSTSMVTASLFNIGLAVRR379 pKa = 11.84DD380 pKa = 3.67SMLRR384 pKa = 11.84GIANDD389 pKa = 3.58SLSGVDD395 pKa = 3.37TSLFQQPEE403 pKa = 4.44LAWAAAVSLACGIDD417 pKa = 3.47VPLNGMSNCYY427 pKa = 10.04VYY429 pKa = 11.15YY430 pKa = 8.46PTLADD435 pKa = 3.68SNVRR439 pKa = 11.84LALPALIKK447 pKa = 9.86TPRR450 pKa = 11.84GYY452 pKa = 9.89WIEE455 pKa = 4.11GDD457 pKa = 3.96SIVIDD462 pKa = 3.55GVPFVGSPYY471 pKa = 10.62VLYY474 pKa = 10.02PLAVYY479 pKa = 10.45DD480 pKa = 4.11FANPYY485 pKa = 10.02SGAFALPKK493 pKa = 9.79PIRR496 pKa = 11.84RR497 pKa = 11.84TRR499 pKa = 11.84KK500 pKa = 7.97GAVFTVIGAWMYY512 pKa = 10.84SWVARR517 pKa = 11.84LAGYY521 pKa = 9.44DD522 pKa = 3.15VHH524 pKa = 9.03ISVNGSNVDD533 pKa = 3.42YY534 pKa = 11.1YY535 pKa = 11.46KK536 pKa = 11.17YY537 pKa = 10.31FASNEE542 pKa = 4.06NSWVHH547 pKa = 5.6PLWNGISDD555 pKa = 4.03EE556 pKa = 4.24IEE558 pKa = 4.11GVFVHH563 pKa = 7.25AIVRR567 pKa = 11.84RR568 pKa = 11.84NKK570 pKa = 10.2HH571 pKa = 5.78FLDD574 pKa = 3.85IPDD577 pKa = 3.43YY578 pKa = 9.86TIPGNLTDD586 pKa = 3.43VSVSVKK592 pKa = 10.33ILDD595 pKa = 3.95RR596 pKa = 11.84YY597 pKa = 10.69VSDD600 pKa = 4.16SSLTTKK606 pKa = 10.21FRR608 pKa = 11.84ANYY611 pKa = 9.2NVGSVTVPKK620 pKa = 10.61SGIQVVTSGDD630 pKa = 3.31LRR632 pKa = 11.84KK633 pKa = 9.37FWGYY637 pKa = 7.85VRR639 pKa = 11.84RR640 pKa = 11.84SQPGLTMVGMAQPAIMPQPKK660 pKa = 9.05HH661 pKa = 5.55QLVEE665 pKa = 3.95IDD667 pKa = 3.88QEE669 pKa = 4.42PEE671 pKa = 3.84VTSEE675 pKa = 3.92MVNMIEE681 pKa = 4.33
Molecular weight: 76.21 kDa
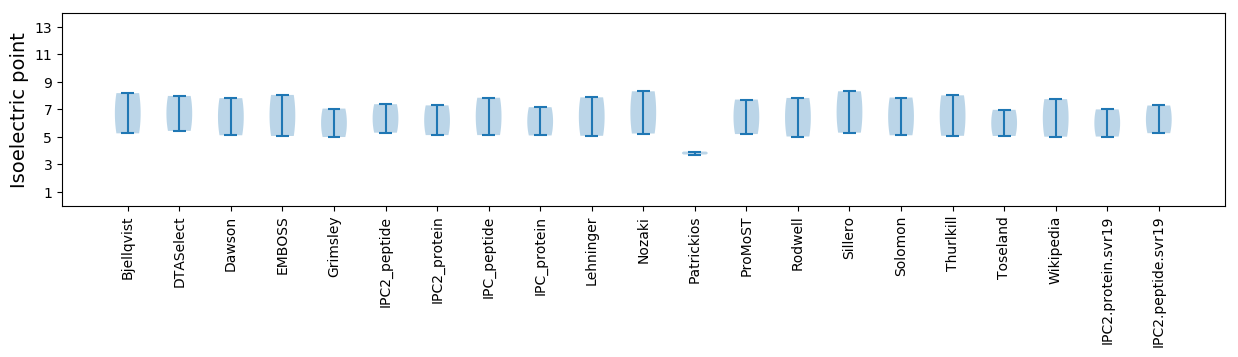
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E5KIN7|E5KIN7_9VIRU RNA-directed RNA polymerase OS=Tuber aestivum virus 1 OX=927810 PE=3 SV=1
MM1 pKa = 7.37CAVSRR6 pKa = 11.84HH7 pKa = 5.66SSTHH11 pKa = 4.91MLVDD15 pKa = 5.21VIPCSQLGLGAVYY28 pKa = 10.25YY29 pKa = 8.48HH30 pKa = 6.44TEE32 pKa = 3.81VAVRR36 pKa = 11.84VYY38 pKa = 11.17GEE40 pKa = 3.75ARR42 pKa = 11.84YY43 pKa = 9.81CRR45 pKa = 11.84LFRR48 pKa = 11.84LDD50 pKa = 5.23DD51 pKa = 3.77VYY53 pKa = 11.53GCYY56 pKa = 9.58MRR58 pKa = 11.84TDD60 pKa = 3.53AYY62 pKa = 9.19TPCLGPVTKK71 pKa = 10.26RR72 pKa = 11.84LMSQVYY78 pKa = 9.6SLVNSVCFNSCDD90 pKa = 3.25ISDD93 pKa = 4.13YY94 pKa = 11.52LSLTLGSTLQMPKK107 pKa = 9.65IEE109 pKa = 4.31WPVKK113 pKa = 8.65HH114 pKa = 6.62GIYY117 pKa = 10.04EE118 pKa = 4.09RR119 pKa = 11.84SKK121 pKa = 9.47ITATHH126 pKa = 5.88HH127 pKa = 4.55THH129 pKa = 7.23LRR131 pKa = 11.84PQEE134 pKa = 4.03LEE136 pKa = 3.96AVAEE140 pKa = 4.39GMGGRR145 pKa = 11.84TKK147 pKa = 10.45FVYY150 pKa = 9.95AVCLQRR156 pKa = 11.84LLSIGVVTEE165 pKa = 4.08AVYY168 pKa = 11.06SSFLAYY174 pKa = 9.81IIGVEE179 pKa = 4.17YY180 pKa = 11.08NFALLLLRR188 pKa = 11.84STTVWSGTRR197 pKa = 11.84GEE199 pKa = 4.15EE200 pKa = 4.04QLLSKK205 pKa = 10.73LKK207 pKa = 10.82SFASAIKK214 pKa = 9.87ASHH217 pKa = 5.18SAKK220 pKa = 10.21YY221 pKa = 10.01QPLTQLFEE229 pKa = 4.31LTVLLNRR236 pKa = 11.84GIGSVDD242 pKa = 2.76WAGEE246 pKa = 3.82KK247 pKa = 10.29EE248 pKa = 4.16NRR250 pKa = 11.84VNPNTVDD257 pKa = 4.32LDD259 pKa = 3.75PADD262 pKa = 3.97VYY264 pKa = 10.8KK265 pKa = 11.04SSMTIFRR272 pKa = 11.84KK273 pKa = 9.64ARR275 pKa = 11.84KK276 pKa = 9.21SGYY279 pKa = 9.44KK280 pKa = 8.41YY281 pKa = 10.94PKK283 pKa = 7.81MHH285 pKa = 6.63YY286 pKa = 10.35DD287 pKa = 3.15KK288 pKa = 10.75FIEE291 pKa = 5.14SRR293 pKa = 11.84WEE295 pKa = 3.96WIPGGSVHH303 pKa = 6.1SQYY306 pKa = 11.16VDD308 pKa = 3.41DD309 pKa = 6.16DD310 pKa = 4.35EE311 pKa = 5.43YY312 pKa = 10.73IYY314 pKa = 10.64PGLYY318 pKa = 8.89TRR320 pKa = 11.84NKK322 pKa = 9.6FITVNKK328 pKa = 8.8MPSHH332 pKa = 6.73HH333 pKa = 6.68LQLFLSSTPQIKK345 pKa = 9.9AWTSTKK351 pKa = 10.5YY352 pKa = 10.25EE353 pKa = 3.85WGKK356 pKa = 9.95QRR358 pKa = 11.84AIYY361 pKa = 8.59GTDD364 pKa = 3.24LRR366 pKa = 11.84STIITNFAMYY376 pKa = 9.99RR377 pKa = 11.84CEE379 pKa = 4.29EE380 pKa = 4.21VLLHH384 pKa = 6.45LFPVGDD390 pKa = 3.53QADD393 pKa = 3.74VEE395 pKa = 4.61KK396 pKa = 10.39VHH398 pKa = 6.81RR399 pKa = 11.84RR400 pKa = 11.84ISMMLDD406 pKa = 3.65GYY408 pKa = 11.41DD409 pKa = 4.04SFCFDD414 pKa = 3.66YY415 pKa = 11.4DD416 pKa = 4.03DD417 pKa = 5.69FNSQHH422 pKa = 6.44SLSSMQAVLLAYY434 pKa = 9.34MDD436 pKa = 4.26AFSDD440 pKa = 3.95VMSPEE445 pKa = 3.89QLQAMEE451 pKa = 4.12WVILSVADD459 pKa = 3.84MQALSPTDD467 pKa = 3.63NEE469 pKa = 4.48WYY471 pKa = 10.41RR472 pKa = 11.84LTGTLLSGWRR482 pKa = 11.84LTTFMNTVLNWVYY495 pKa = 10.39MDD497 pKa = 3.33VAGVFNIDD505 pKa = 3.41GVHH508 pKa = 6.93DD509 pKa = 4.24SVHH512 pKa = 6.52NGDD515 pKa = 4.81DD516 pKa = 3.66VMISISSIKK525 pKa = 10.15AANLVMNKK533 pKa = 6.24MHH535 pKa = 6.9KK536 pKa = 10.34VNARR540 pKa = 11.84AQPTKK545 pKa = 10.81CNVLSISEE553 pKa = 4.07FLRR556 pKa = 11.84IEE558 pKa = 4.29HH559 pKa = 6.23GMSGYY564 pKa = 10.74DD565 pKa = 3.52GLGAQYY571 pKa = 10.96LSRR574 pKa = 11.84SCATLVHH581 pKa = 6.22SRR583 pKa = 11.84VEE585 pKa = 4.22SKK587 pKa = 11.1EE588 pKa = 4.08PITATRR594 pKa = 11.84LVEE597 pKa = 4.44ADD599 pKa = 3.77LSRR602 pKa = 11.84LKK604 pKa = 10.54DD605 pKa = 3.61YY606 pKa = 10.94RR607 pKa = 11.84SRR609 pKa = 11.84CANVDD614 pKa = 3.47AVDD617 pKa = 6.02LIQQQLFCRR626 pKa = 11.84TAKK629 pKa = 10.44LFNIEE634 pKa = 4.52LGNIYY639 pKa = 10.03KK640 pKa = 10.06ISEE643 pKa = 3.97LHH645 pKa = 6.06RR646 pKa = 11.84VVGGCSDD653 pKa = 3.89RR654 pKa = 11.84RR655 pKa = 11.84WAPIEE660 pKa = 3.93QQVTTTSGKK669 pKa = 10.03YY670 pKa = 9.13EE671 pKa = 4.15IPHH674 pKa = 6.65EE675 pKa = 4.4IDD677 pKa = 5.73DD678 pKa = 4.07PSFWPGVNDD687 pKa = 3.38YY688 pKa = 11.57SAMLYY693 pKa = 10.31RR694 pKa = 11.84KK695 pKa = 9.64IGEE698 pKa = 4.14QVKK701 pKa = 9.84MSVIVDD707 pKa = 3.81AVSQGSRR714 pKa = 11.84LTIAKK719 pKa = 9.7SRR721 pKa = 11.84SVTLAVEE728 pKa = 4.5RR729 pKa = 11.84TANIKK734 pKa = 9.95AKK736 pKa = 8.27EE737 pKa = 4.15WEE739 pKa = 3.92RR740 pKa = 11.84ALYY743 pKa = 8.33RR744 pKa = 11.84TYY746 pKa = 10.79KK747 pKa = 10.48GSTVHH752 pKa = 6.48YY753 pKa = 8.73YY754 pKa = 11.51ANLAKK759 pKa = 10.15FLNVPPLFGIEE770 pKa = 4.13TRR772 pKa = 11.84GGASAISAALASSDD786 pKa = 3.15TMRR789 pKa = 11.84ALKK792 pKa = 10.69VLLL795 pKa = 4.21
MM1 pKa = 7.37CAVSRR6 pKa = 11.84HH7 pKa = 5.66SSTHH11 pKa = 4.91MLVDD15 pKa = 5.21VIPCSQLGLGAVYY28 pKa = 10.25YY29 pKa = 8.48HH30 pKa = 6.44TEE32 pKa = 3.81VAVRR36 pKa = 11.84VYY38 pKa = 11.17GEE40 pKa = 3.75ARR42 pKa = 11.84YY43 pKa = 9.81CRR45 pKa = 11.84LFRR48 pKa = 11.84LDD50 pKa = 5.23DD51 pKa = 3.77VYY53 pKa = 11.53GCYY56 pKa = 9.58MRR58 pKa = 11.84TDD60 pKa = 3.53AYY62 pKa = 9.19TPCLGPVTKK71 pKa = 10.26RR72 pKa = 11.84LMSQVYY78 pKa = 9.6SLVNSVCFNSCDD90 pKa = 3.25ISDD93 pKa = 4.13YY94 pKa = 11.52LSLTLGSTLQMPKK107 pKa = 9.65IEE109 pKa = 4.31WPVKK113 pKa = 8.65HH114 pKa = 6.62GIYY117 pKa = 10.04EE118 pKa = 4.09RR119 pKa = 11.84SKK121 pKa = 9.47ITATHH126 pKa = 5.88HH127 pKa = 4.55THH129 pKa = 7.23LRR131 pKa = 11.84PQEE134 pKa = 4.03LEE136 pKa = 3.96AVAEE140 pKa = 4.39GMGGRR145 pKa = 11.84TKK147 pKa = 10.45FVYY150 pKa = 9.95AVCLQRR156 pKa = 11.84LLSIGVVTEE165 pKa = 4.08AVYY168 pKa = 11.06SSFLAYY174 pKa = 9.81IIGVEE179 pKa = 4.17YY180 pKa = 11.08NFALLLLRR188 pKa = 11.84STTVWSGTRR197 pKa = 11.84GEE199 pKa = 4.15EE200 pKa = 4.04QLLSKK205 pKa = 10.73LKK207 pKa = 10.82SFASAIKK214 pKa = 9.87ASHH217 pKa = 5.18SAKK220 pKa = 10.21YY221 pKa = 10.01QPLTQLFEE229 pKa = 4.31LTVLLNRR236 pKa = 11.84GIGSVDD242 pKa = 2.76WAGEE246 pKa = 3.82KK247 pKa = 10.29EE248 pKa = 4.16NRR250 pKa = 11.84VNPNTVDD257 pKa = 4.32LDD259 pKa = 3.75PADD262 pKa = 3.97VYY264 pKa = 10.8KK265 pKa = 11.04SSMTIFRR272 pKa = 11.84KK273 pKa = 9.64ARR275 pKa = 11.84KK276 pKa = 9.21SGYY279 pKa = 9.44KK280 pKa = 8.41YY281 pKa = 10.94PKK283 pKa = 7.81MHH285 pKa = 6.63YY286 pKa = 10.35DD287 pKa = 3.15KK288 pKa = 10.75FIEE291 pKa = 5.14SRR293 pKa = 11.84WEE295 pKa = 3.96WIPGGSVHH303 pKa = 6.1SQYY306 pKa = 11.16VDD308 pKa = 3.41DD309 pKa = 6.16DD310 pKa = 4.35EE311 pKa = 5.43YY312 pKa = 10.73IYY314 pKa = 10.64PGLYY318 pKa = 8.89TRR320 pKa = 11.84NKK322 pKa = 9.6FITVNKK328 pKa = 8.8MPSHH332 pKa = 6.73HH333 pKa = 6.68LQLFLSSTPQIKK345 pKa = 9.9AWTSTKK351 pKa = 10.5YY352 pKa = 10.25EE353 pKa = 3.85WGKK356 pKa = 9.95QRR358 pKa = 11.84AIYY361 pKa = 8.59GTDD364 pKa = 3.24LRR366 pKa = 11.84STIITNFAMYY376 pKa = 9.99RR377 pKa = 11.84CEE379 pKa = 4.29EE380 pKa = 4.21VLLHH384 pKa = 6.45LFPVGDD390 pKa = 3.53QADD393 pKa = 3.74VEE395 pKa = 4.61KK396 pKa = 10.39VHH398 pKa = 6.81RR399 pKa = 11.84RR400 pKa = 11.84ISMMLDD406 pKa = 3.65GYY408 pKa = 11.41DD409 pKa = 4.04SFCFDD414 pKa = 3.66YY415 pKa = 11.4DD416 pKa = 4.03DD417 pKa = 5.69FNSQHH422 pKa = 6.44SLSSMQAVLLAYY434 pKa = 9.34MDD436 pKa = 4.26AFSDD440 pKa = 3.95VMSPEE445 pKa = 3.89QLQAMEE451 pKa = 4.12WVILSVADD459 pKa = 3.84MQALSPTDD467 pKa = 3.63NEE469 pKa = 4.48WYY471 pKa = 10.41RR472 pKa = 11.84LTGTLLSGWRR482 pKa = 11.84LTTFMNTVLNWVYY495 pKa = 10.39MDD497 pKa = 3.33VAGVFNIDD505 pKa = 3.41GVHH508 pKa = 6.93DD509 pKa = 4.24SVHH512 pKa = 6.52NGDD515 pKa = 4.81DD516 pKa = 3.66VMISISSIKK525 pKa = 10.15AANLVMNKK533 pKa = 6.24MHH535 pKa = 6.9KK536 pKa = 10.34VNARR540 pKa = 11.84AQPTKK545 pKa = 10.81CNVLSISEE553 pKa = 4.07FLRR556 pKa = 11.84IEE558 pKa = 4.29HH559 pKa = 6.23GMSGYY564 pKa = 10.74DD565 pKa = 3.52GLGAQYY571 pKa = 10.96LSRR574 pKa = 11.84SCATLVHH581 pKa = 6.22SRR583 pKa = 11.84VEE585 pKa = 4.22SKK587 pKa = 11.1EE588 pKa = 4.08PITATRR594 pKa = 11.84LVEE597 pKa = 4.44ADD599 pKa = 3.77LSRR602 pKa = 11.84LKK604 pKa = 10.54DD605 pKa = 3.61YY606 pKa = 10.94RR607 pKa = 11.84SRR609 pKa = 11.84CANVDD614 pKa = 3.47AVDD617 pKa = 6.02LIQQQLFCRR626 pKa = 11.84TAKK629 pKa = 10.44LFNIEE634 pKa = 4.52LGNIYY639 pKa = 10.03KK640 pKa = 10.06ISEE643 pKa = 3.97LHH645 pKa = 6.06RR646 pKa = 11.84VVGGCSDD653 pKa = 3.89RR654 pKa = 11.84RR655 pKa = 11.84WAPIEE660 pKa = 3.93QQVTTTSGKK669 pKa = 10.03YY670 pKa = 9.13EE671 pKa = 4.15IPHH674 pKa = 6.65EE675 pKa = 4.4IDD677 pKa = 5.73DD678 pKa = 4.07PSFWPGVNDD687 pKa = 3.38YY688 pKa = 11.57SAMLYY693 pKa = 10.31RR694 pKa = 11.84KK695 pKa = 9.64IGEE698 pKa = 4.14QVKK701 pKa = 9.84MSVIVDD707 pKa = 3.81AVSQGSRR714 pKa = 11.84LTIAKK719 pKa = 9.7SRR721 pKa = 11.84SVTLAVEE728 pKa = 4.5RR729 pKa = 11.84TANIKK734 pKa = 9.95AKK736 pKa = 8.27EE737 pKa = 4.15WEE739 pKa = 3.92RR740 pKa = 11.84ALYY743 pKa = 8.33RR744 pKa = 11.84TYY746 pKa = 10.79KK747 pKa = 10.48GSTVHH752 pKa = 6.48YY753 pKa = 8.73YY754 pKa = 11.51ANLAKK759 pKa = 10.15FLNVPPLFGIEE770 pKa = 4.13TRR772 pKa = 11.84GGASAISAALASSDD786 pKa = 3.15TMRR789 pKa = 11.84ALKK792 pKa = 10.69VLLL795 pKa = 4.21
Molecular weight: 89.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1476 |
681 |
795 |
738.0 |
83.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.182 ± 0.008 | 1.152 ± 0.52 |
5.488 ± 0.145 | 4.607 ± 0.211 |
3.388 ± 0.172 | 6.098 ± 0.309 |
2.168 ± 0.513 | 5.285 ± 0.09 |
4.336 ± 0.403 | 9.824 ± 0.187 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.049 ± 0.157 | 4.675 ± 0.904 |
4.404 ± 0.891 | 3.117 ± 0.198 |
5.081 ± 0.499 | 8.401 ± 0.553 |
5.962 ± 0.054 | 8.333 ± 0.111 |
2.168 ± 0.288 | 5.285 ± 0.001 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
